Manganese »
PDB 9c9w-9j1a »
9cag »
Manganese in PDB 9cag: Human TOP3B-TDRD3 Core Complex in Rna Pre-Cleavage State
Enzymatic activity of Human TOP3B-TDRD3 Core Complex in Rna Pre-Cleavage State
All present enzymatic activity of Human TOP3B-TDRD3 Core Complex in Rna Pre-Cleavage State:
5.6.2.1;
5.6.2.1;
Manganese Binding Sites:
The binding sites of Manganese atom in the Human TOP3B-TDRD3 Core Complex in Rna Pre-Cleavage State
(pdb code 9cag). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 2 binding sites of Manganese where determined in the Human TOP3B-TDRD3 Core Complex in Rna Pre-Cleavage State, PDB code: 9cag:
Jump to Manganese binding site number: 1; 2;
In total 2 binding sites of Manganese where determined in the Human TOP3B-TDRD3 Core Complex in Rna Pre-Cleavage State, PDB code: 9cag:
Jump to Manganese binding site number: 1; 2;
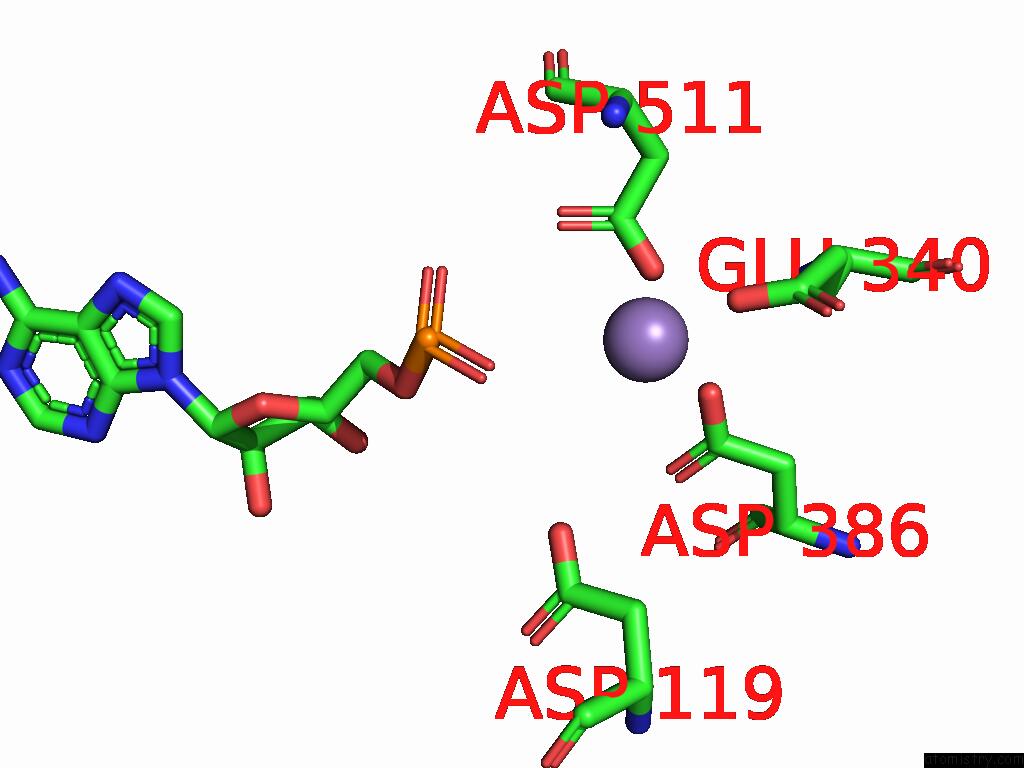
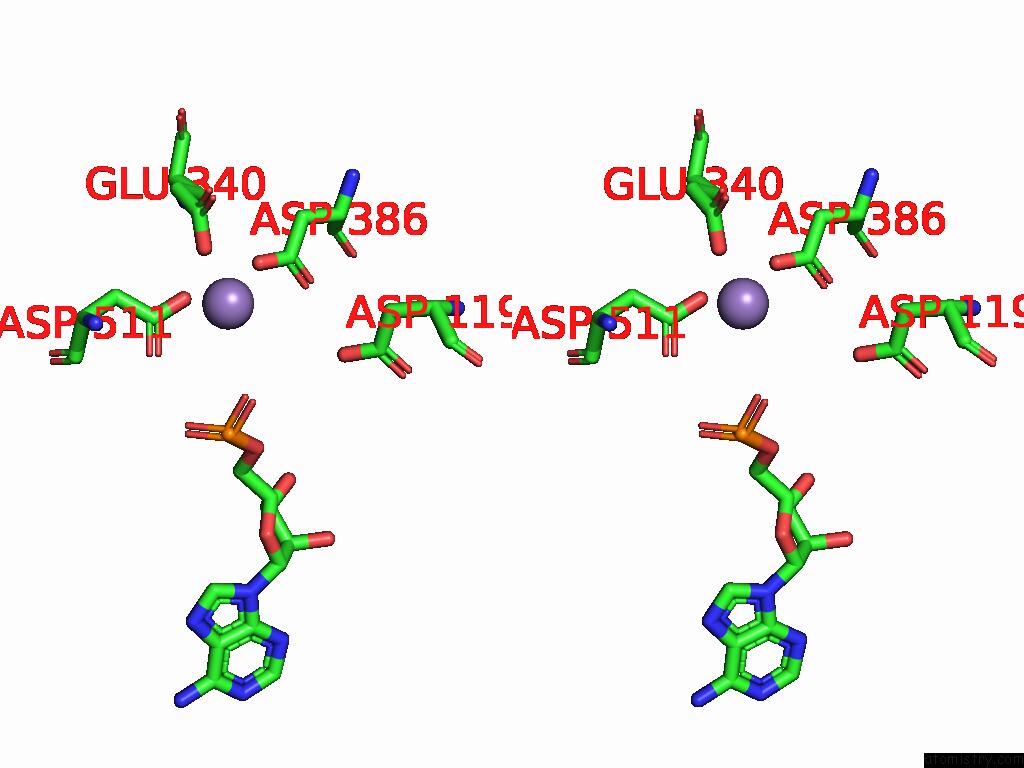
Manganese binding site 1 out of 2 in 9cag
Go back to
Manganese binding site 1 out
of 2 in the Human TOP3B-TDRD3 Core Complex in Rna Pre-Cleavage State
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Human TOP3B-TDRD3 Core Complex in Rna Pre-Cleavage State within 5.0Å range:
|
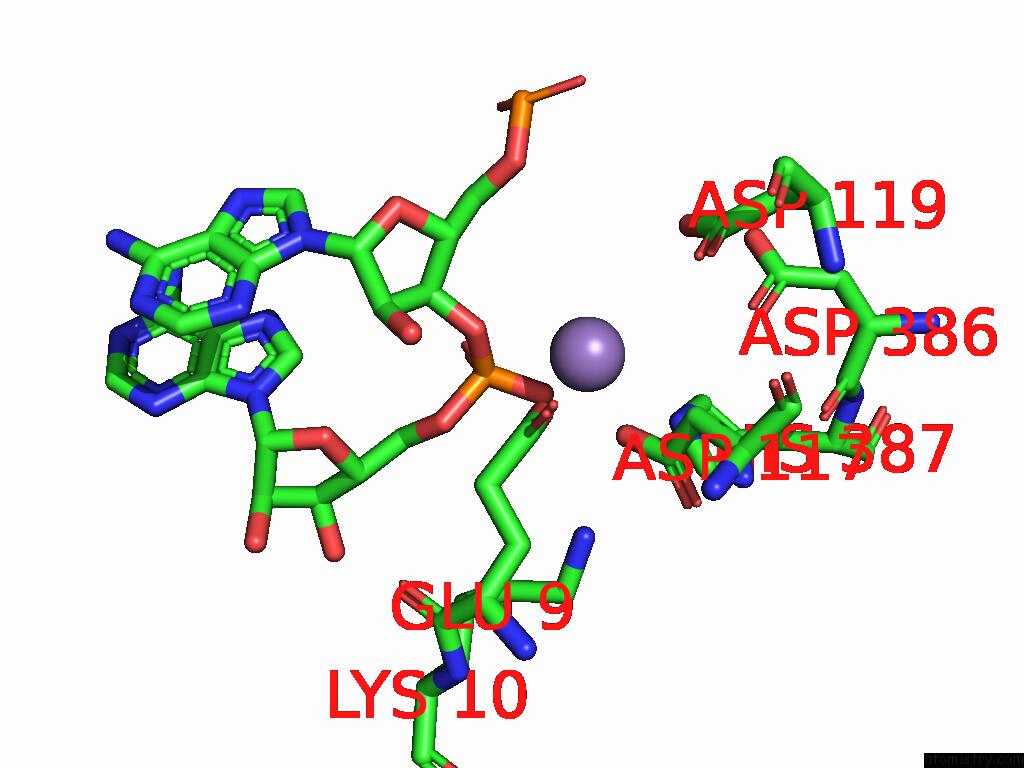
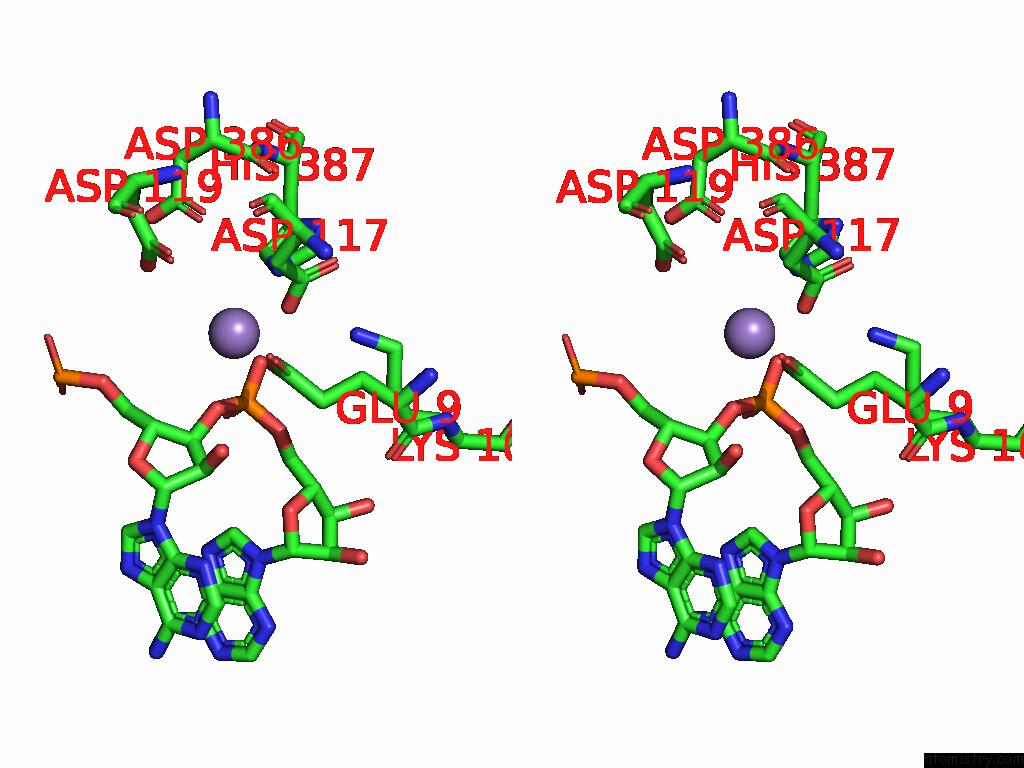
Manganese binding site 2 out of 2 in 9cag
Go back to
Manganese binding site 2 out
of 2 in the Human TOP3B-TDRD3 Core Complex in Rna Pre-Cleavage State
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Human TOP3B-TDRD3 Core Complex in Rna Pre-Cleavage State within 5.0Å range:
|
Reference:
X.Yang,
X.Chen,
W.Yang,
Y.Pommier.
Structural Insights Into Human Topoisomerase 3 Beta Dna and Rna Catalysis and Nucleic Acid Gate Dynamics. Nat Commun V. 16 834 2025.
ISSN: ESSN 2041-1723
PubMed: 39828754
DOI: 10.1038/S41467-025-55959-Y
Page generated: Sun Feb 9 08:28:30 2025
ISSN: ESSN 2041-1723
PubMed: 39828754
DOI: 10.1038/S41467-025-55959-Y
Last articles
K in 8K0TK in 8JGW
K in 8JZA
K in 8JC7
K in 8JGR
K in 8IYM
K in 8JAH
K in 8IAX
K in 8IPP
K in 8IBN