Manganese »
PDB 8zge-9bya »
9bx2 »
Manganese in PDB 9bx2: Class 2 Model For Preturnover Condition of Bacillus Subtilis Ribonucleotide Reductase Complex
Enzymatic activity of Class 2 Model For Preturnover Condition of Bacillus Subtilis Ribonucleotide Reductase Complex
All present enzymatic activity of Class 2 Model For Preturnover Condition of Bacillus Subtilis Ribonucleotide Reductase Complex:
1.17.4.1;
1.17.4.1;
Other elements in 9bx2:
The structure of Class 2 Model For Preturnover Condition of Bacillus Subtilis Ribonucleotide Reductase Complex also contains other interesting chemical elements:
| Magnesium | (Mg) | 2 atoms |
Manganese Binding Sites:
The binding sites of Manganese atom in the Class 2 Model For Preturnover Condition of Bacillus Subtilis Ribonucleotide Reductase Complex
(pdb code 9bx2). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 4 binding sites of Manganese where determined in the Class 2 Model For Preturnover Condition of Bacillus Subtilis Ribonucleotide Reductase Complex, PDB code: 9bx2:
Jump to Manganese binding site number: 1; 2; 3; 4;
In total 4 binding sites of Manganese where determined in the Class 2 Model For Preturnover Condition of Bacillus Subtilis Ribonucleotide Reductase Complex, PDB code: 9bx2:
Jump to Manganese binding site number: 1; 2; 3; 4;
Manganese binding site 1 out of 4 in 9bx2
Go back to
Manganese binding site 1 out
of 4 in the Class 2 Model For Preturnover Condition of Bacillus Subtilis Ribonucleotide Reductase Complex
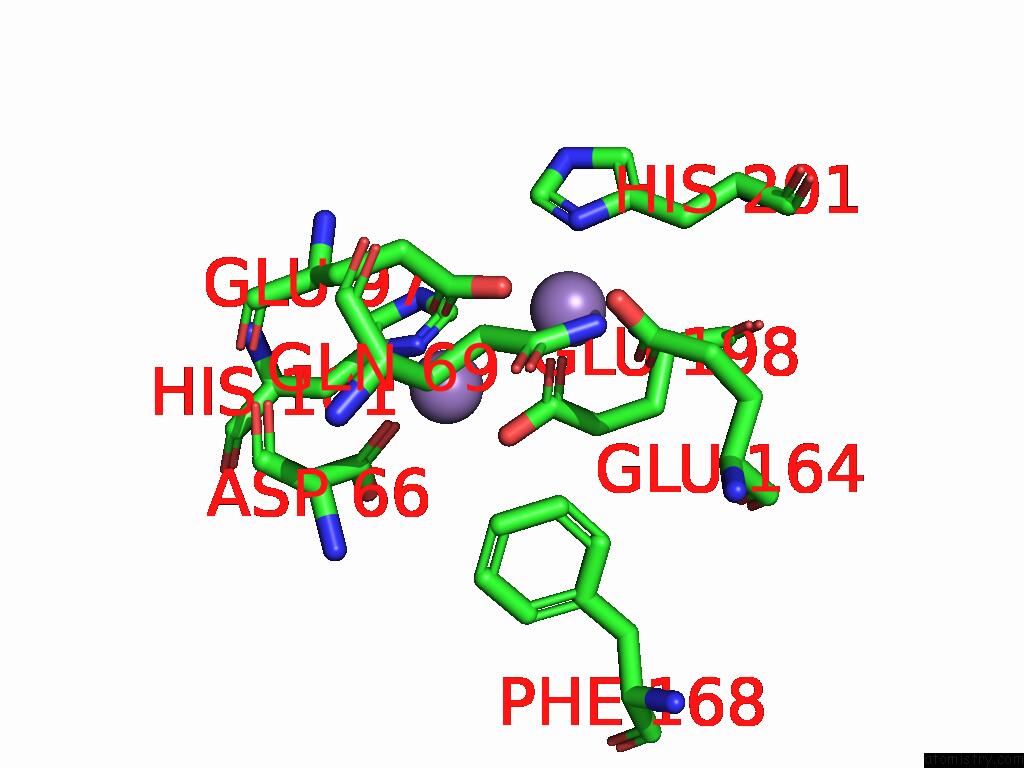
Mono view
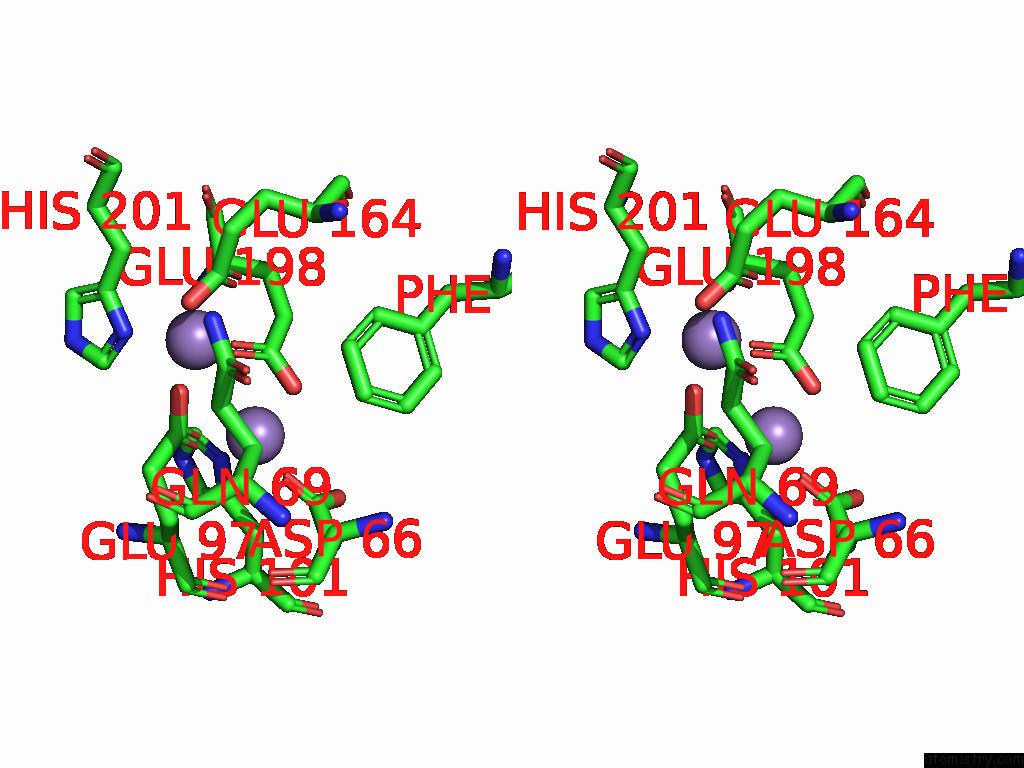
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Class 2 Model For Preturnover Condition of Bacillus Subtilis Ribonucleotide Reductase Complex within 5.0Å range:
|
Manganese binding site 2 out of 4 in 9bx2
Go back to
Manganese binding site 2 out
of 4 in the Class 2 Model For Preturnover Condition of Bacillus Subtilis Ribonucleotide Reductase Complex
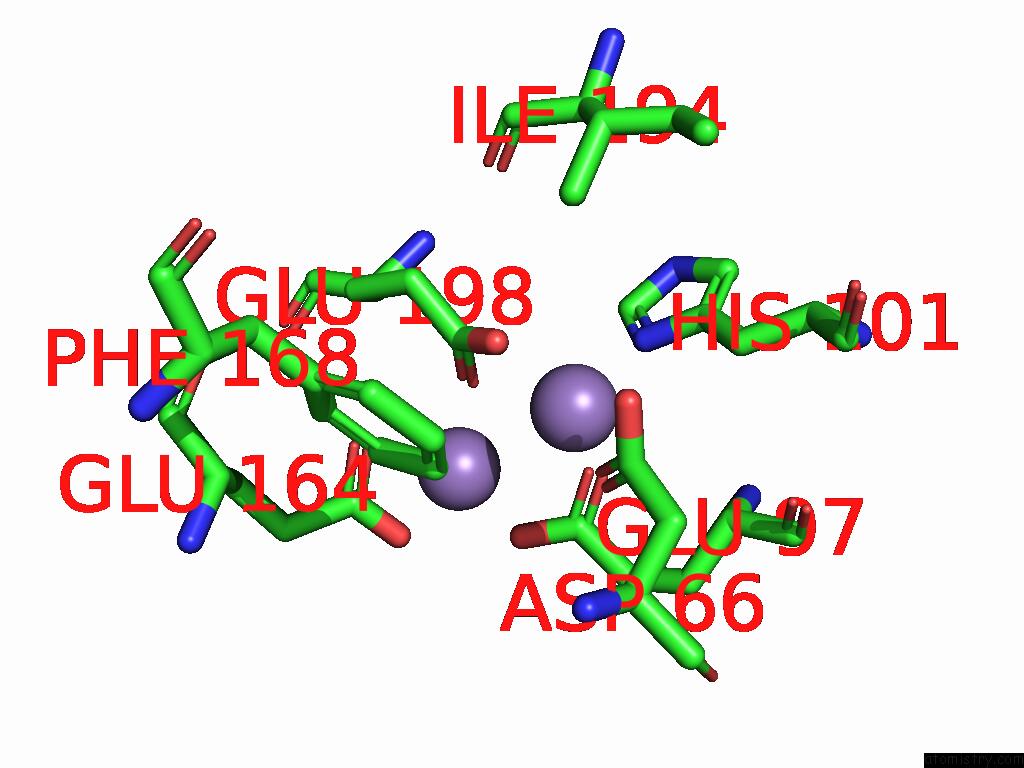
Mono view
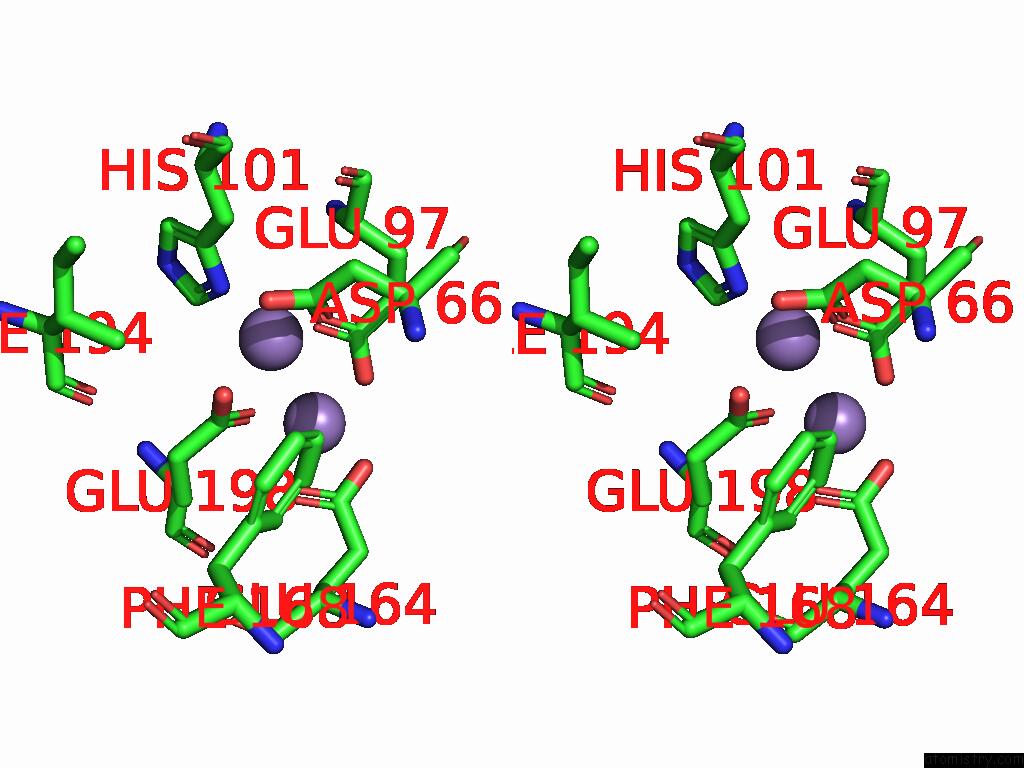
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Class 2 Model For Preturnover Condition of Bacillus Subtilis Ribonucleotide Reductase Complex within 5.0Å range:
|
Manganese binding site 3 out of 4 in 9bx2
Go back to
Manganese binding site 3 out
of 4 in the Class 2 Model For Preturnover Condition of Bacillus Subtilis Ribonucleotide Reductase Complex
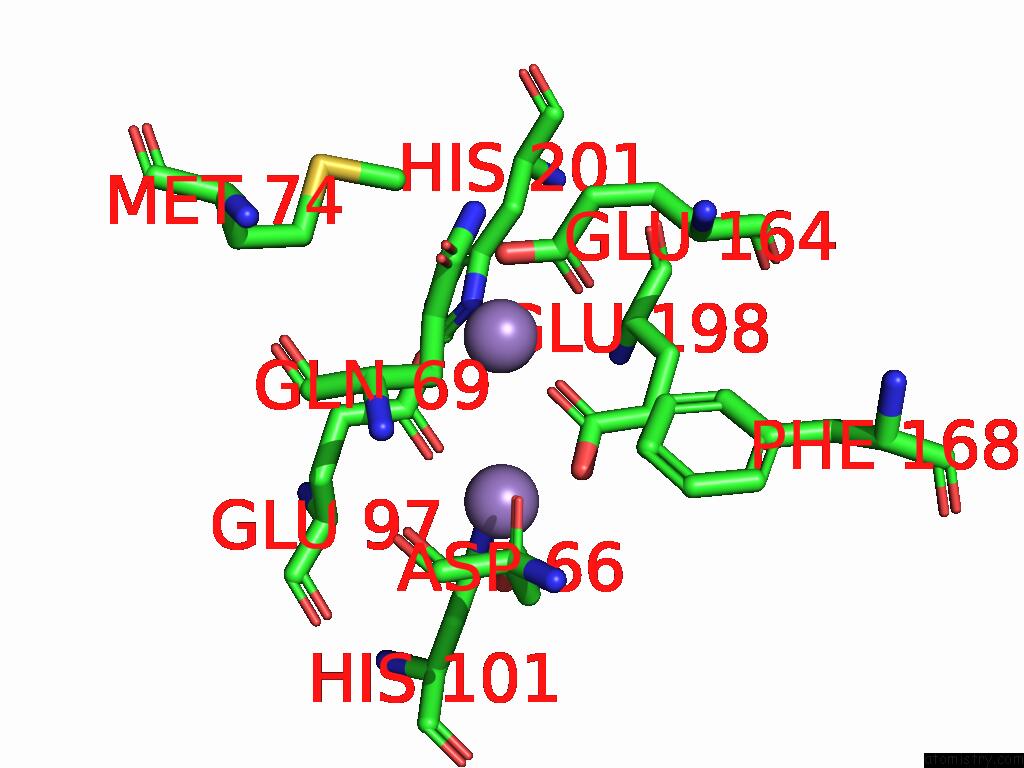
Mono view
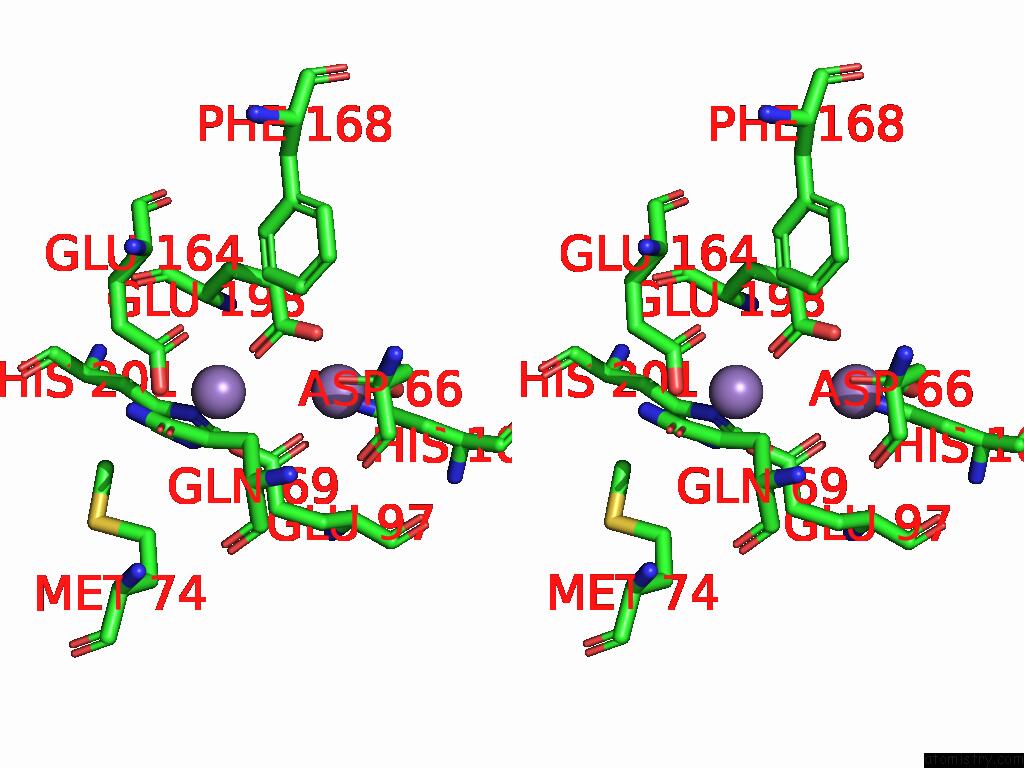
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Class 2 Model For Preturnover Condition of Bacillus Subtilis Ribonucleotide Reductase Complex within 5.0Å range:
|
Manganese binding site 4 out of 4 in 9bx2
Go back to
Manganese binding site 4 out
of 4 in the Class 2 Model For Preturnover Condition of Bacillus Subtilis Ribonucleotide Reductase Complex
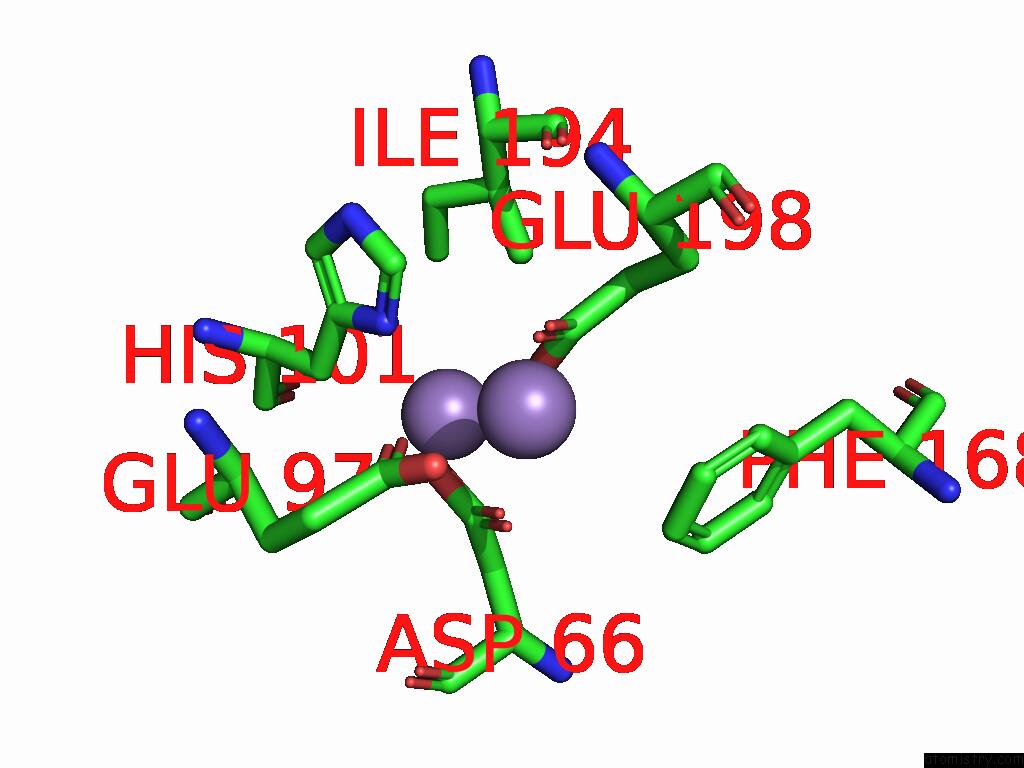
Mono view
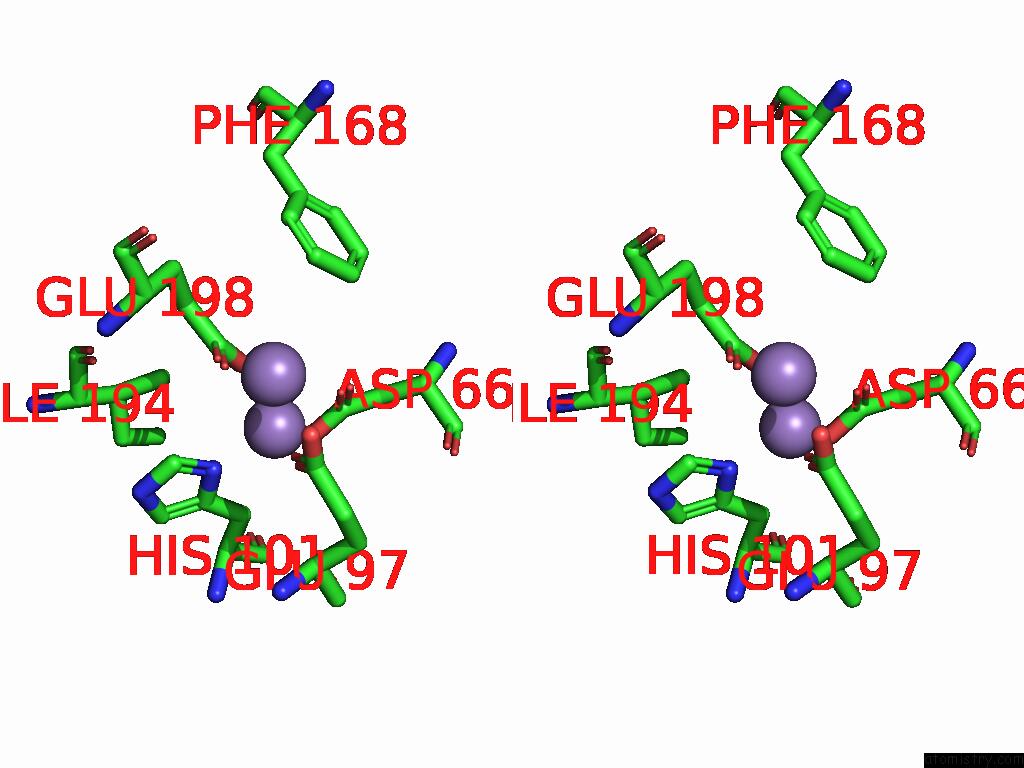
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of Class 2 Model For Preturnover Condition of Bacillus Subtilis Ribonucleotide Reductase Complex within 5.0Å range:
|
Reference:
D.Xu,
W.C.Thomas,
A.A.Burnim,
N.Ando.
Conformational Landscapes of A Class I Ribonucleotide Reductase Complex During Turnover Reveal Intrinsic Dynamics and Asymmetry Nat Commun V. 16 2458 2025.
ISSN: ESSN 2041-1723
DOI: 10.1038/S41467-025-57735-4
Page generated: Sun Aug 17 02:11:10 2025
ISSN: ESSN 2041-1723
DOI: 10.1038/S41467-025-57735-4
Last articles
Na in 2A2CNa in 2A1D
Na in 2A1E
Na in 293D
Na in 284D
Na in 292D
Na in 258D
Na in 1ZV8
Na in 246D
Na in 231D