Manganese »
PDB 4php-4qsf »
4qpx »
Manganese in PDB 4qpx: Nv Polymerase Post-Incorporation-Like Complex
Enzymatic activity of Nv Polymerase Post-Incorporation-Like Complex
All present enzymatic activity of Nv Polymerase Post-Incorporation-Like Complex:
2.7.7.48;
2.7.7.48;
Protein crystallography data
The structure of Nv Polymerase Post-Incorporation-Like Complex, PDB code: 4qpx
was solved by
D.F.Zamyatkin,
F.Parra,
P.Grochulski,
K.K.S.Ng,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.70 / 1.86 |
| Space group | P 41 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 81.220, 81.220, 188.130, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.5 / 25.3 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Nv Polymerase Post-Incorporation-Like Complex
(pdb code 4qpx). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 3 binding sites of Manganese where determined in the Nv Polymerase Post-Incorporation-Like Complex, PDB code: 4qpx:
Jump to Manganese binding site number: 1; 2; 3;
In total 3 binding sites of Manganese where determined in the Nv Polymerase Post-Incorporation-Like Complex, PDB code: 4qpx:
Jump to Manganese binding site number: 1; 2; 3;
Manganese binding site 1 out of 3 in 4qpx
Go back to
Manganese binding site 1 out
of 3 in the Nv Polymerase Post-Incorporation-Like Complex
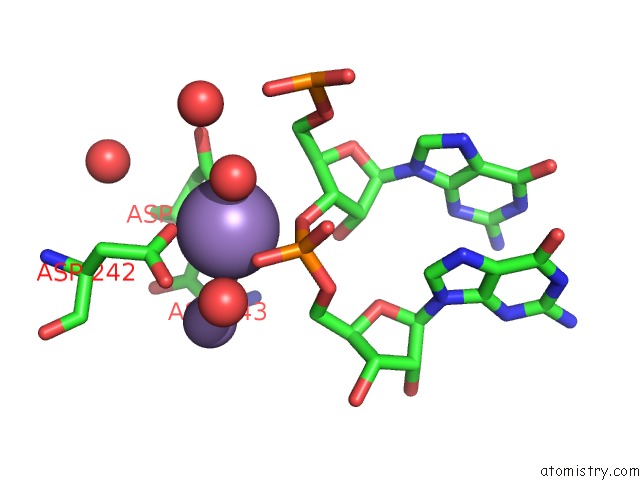
Mono view
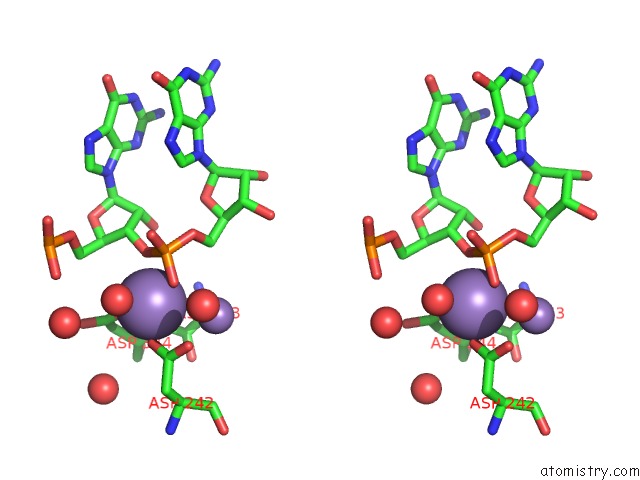
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Nv Polymerase Post-Incorporation-Like Complex within 5.0Å range:
|
Manganese binding site 2 out of 3 in 4qpx
Go back to
Manganese binding site 2 out
of 3 in the Nv Polymerase Post-Incorporation-Like Complex
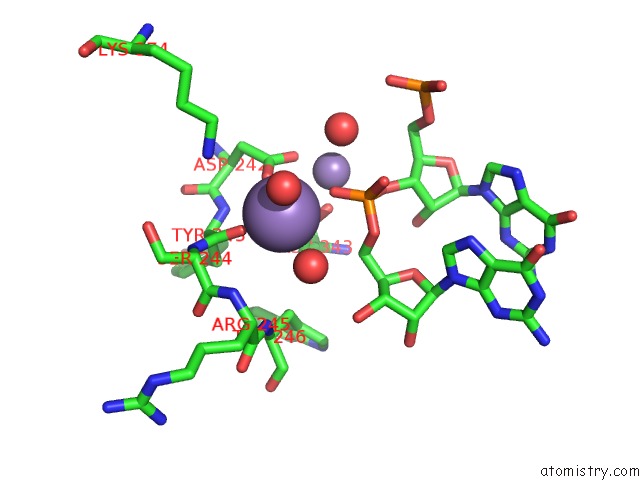
Mono view
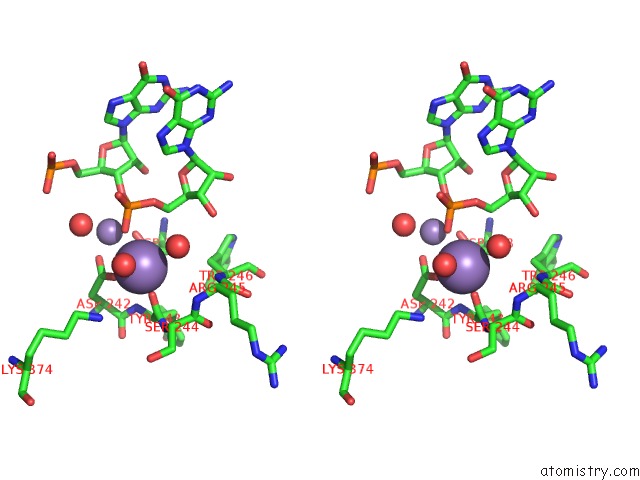
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Nv Polymerase Post-Incorporation-Like Complex within 5.0Å range:
|
Manganese binding site 3 out of 3 in 4qpx
Go back to
Manganese binding site 3 out
of 3 in the Nv Polymerase Post-Incorporation-Like Complex
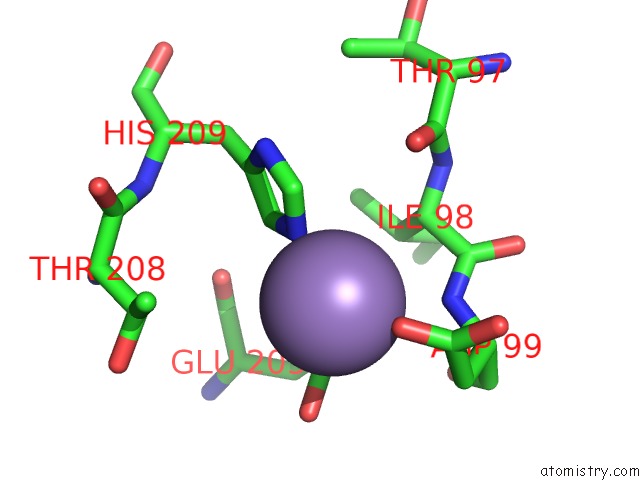
Mono view
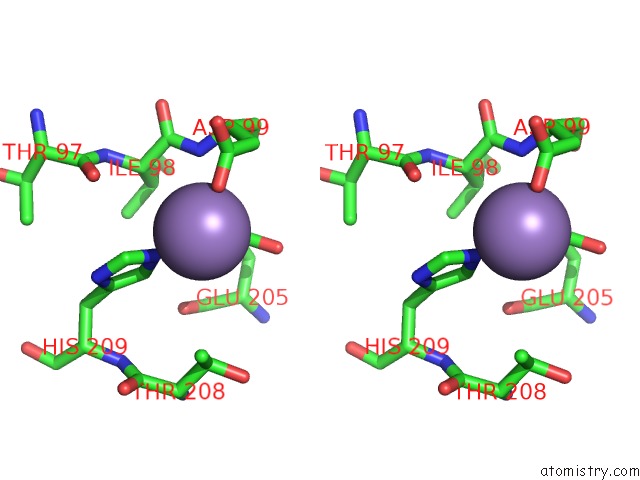
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Nv Polymerase Post-Incorporation-Like Complex within 5.0Å range:
|
Reference:
D.Zamyatkin,
C.Rao,
E.Hoffarth,
G.Jurca,
H.Rho,
F.Parra,
P.Grochulski,
K.K.Ng.
Structure of A Backtracked State Reveals Conformational Changes Similar to the State Following Nucleotide Incorporation in Human Norovirus Polymerase. Acta Crystallogr.,Sect.D V. 70 3099 2014.
ISSN: ISSN 0907-4449
PubMed: 25478829
DOI: 10.1107/S1399004714021518
Page generated: Sat Oct 5 21:01:42 2024
ISSN: ISSN 0907-4449
PubMed: 25478829
DOI: 10.1107/S1399004714021518
Last articles
K in 6FBNK in 6F3N
K in 6F3M
K in 6EVF
K in 6EVE
K in 6F29
K in 6EVV
K in 6EVC
K in 6EVB
K in 6EVD