Manganese »
PDB 7ohl-7sck »
7rpx »
Manganese in PDB 7rpx: Archaeal Dna Ligase and Heterotrimeric Pcna in Complex with End-Joined Dna
Enzymatic activity of Archaeal Dna Ligase and Heterotrimeric Pcna in Complex with End-Joined Dna
All present enzymatic activity of Archaeal Dna Ligase and Heterotrimeric Pcna in Complex with End-Joined Dna:
6.5.1.1;
6.5.1.1;
Manganese Binding Sites:
The binding sites of Manganese atom in the Archaeal Dna Ligase and Heterotrimeric Pcna in Complex with End-Joined Dna
(pdb code 7rpx). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 3 binding sites of Manganese where determined in the Archaeal Dna Ligase and Heterotrimeric Pcna in Complex with End-Joined Dna, PDB code: 7rpx:
Jump to Manganese binding site number: 1; 2; 3;
In total 3 binding sites of Manganese where determined in the Archaeal Dna Ligase and Heterotrimeric Pcna in Complex with End-Joined Dna, PDB code: 7rpx:
Jump to Manganese binding site number: 1; 2; 3;
Manganese binding site 1 out of 3 in 7rpx
Go back to
Manganese binding site 1 out
of 3 in the Archaeal Dna Ligase and Heterotrimeric Pcna in Complex with End-Joined Dna
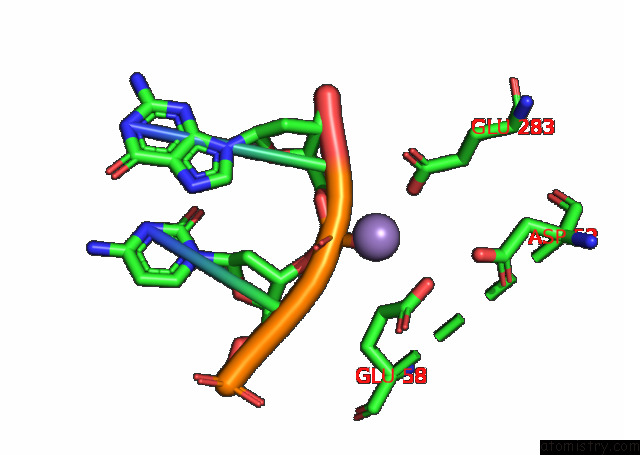
Mono view
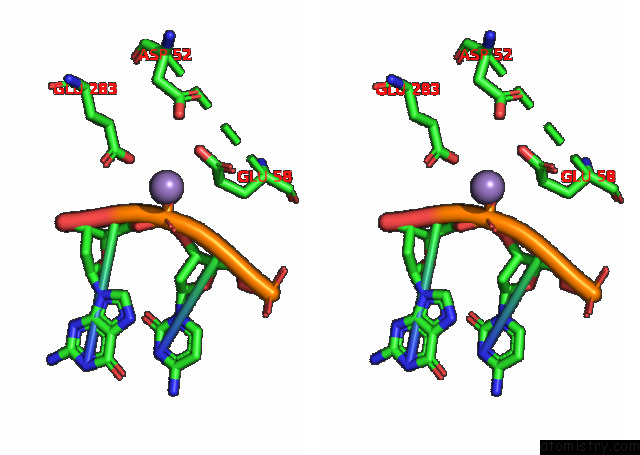
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Archaeal Dna Ligase and Heterotrimeric Pcna in Complex with End-Joined Dna within 5.0Å range:
|
Manganese binding site 2 out of 3 in 7rpx
Go back to
Manganese binding site 2 out
of 3 in the Archaeal Dna Ligase and Heterotrimeric Pcna in Complex with End-Joined Dna
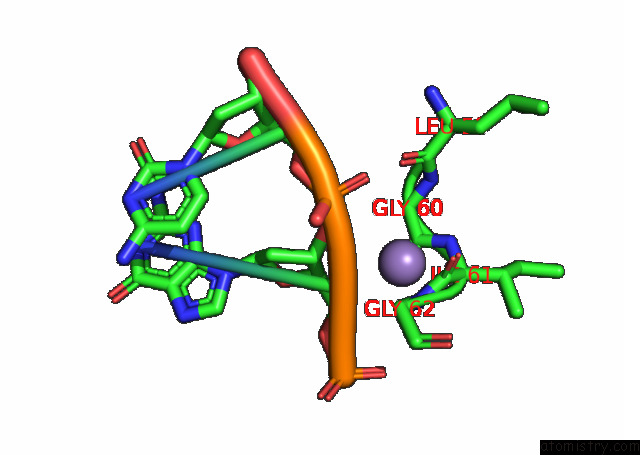
Mono view
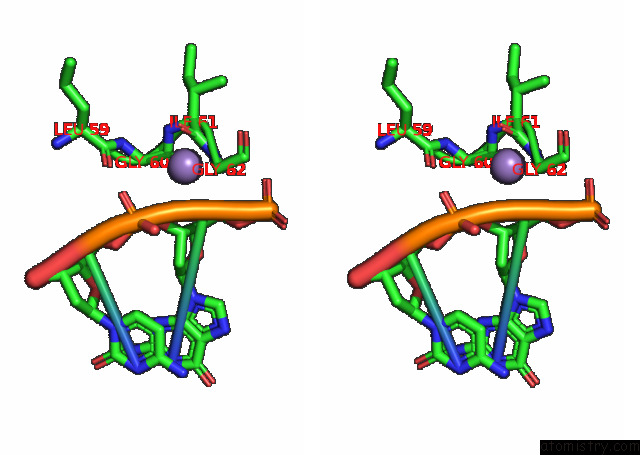
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Archaeal Dna Ligase and Heterotrimeric Pcna in Complex with End-Joined Dna within 5.0Å range:
|
Manganese binding site 3 out of 3 in 7rpx
Go back to
Manganese binding site 3 out
of 3 in the Archaeal Dna Ligase and Heterotrimeric Pcna in Complex with End-Joined Dna
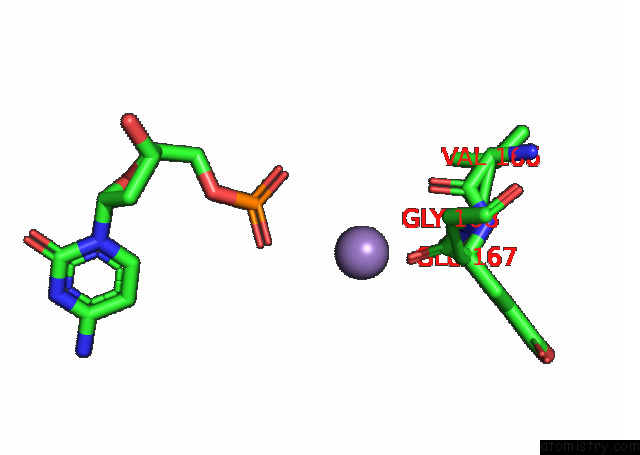
Mono view
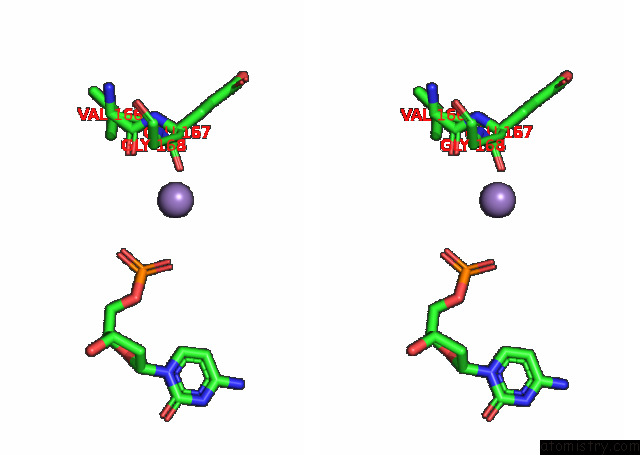
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Archaeal Dna Ligase and Heterotrimeric Pcna in Complex with End-Joined Dna within 5.0Å range:
|
Reference:
A.Sverzhinsky,
A.E.Tomkinson,
J.M.Pascal.
Cryo-Em Structures and Biochemical Insights Into Heterotrimeric Pcna Regulation of Dna Ligase To Be Published.
Page generated: Sat Aug 16 23:35:41 2025
Last articles
Mn in 9ICAMn in 9HDM
Mn in 9G6H
Mn in 9HSH
Mn in 9GSU
Mn in 9H6L
Mn in 9H6K
Mn in 9H6J
Mn in 9G6G
Mn in 9GMW