Manganese »
PDB 9jgm-9xia »
9nb9 »
Manganese in PDB 9nb9: Viral Protein DP71L in Complex with Phosphorylated EIF2ALPHA (Ntd) and Protein Phosphatase 1A (D64A), Stabilized By G-Actin/Dnasei
Enzymatic activity of Viral Protein DP71L in Complex with Phosphorylated EIF2ALPHA (Ntd) and Protein Phosphatase 1A (D64A), Stabilized By G-Actin/Dnasei
All present enzymatic activity of Viral Protein DP71L in Complex with Phosphorylated EIF2ALPHA (Ntd) and Protein Phosphatase 1A (D64A), Stabilized By G-Actin/Dnasei:
3.1.21.1; 3.1.3.16;
3.1.21.1; 3.1.3.16;
Other elements in 9nb9:
The structure of Viral Protein DP71L in Complex with Phosphorylated EIF2ALPHA (Ntd) and Protein Phosphatase 1A (D64A), Stabilized By G-Actin/Dnasei also contains other interesting chemical elements:
| Calcium | (Ca) | 1 atom |
Manganese Binding Sites:
The binding sites of Manganese atom in the Viral Protein DP71L in Complex with Phosphorylated EIF2ALPHA (Ntd) and Protein Phosphatase 1A (D64A), Stabilized By G-Actin/Dnasei
(pdb code 9nb9). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total only one binding site of Manganese was determined in the Viral Protein DP71L in Complex with Phosphorylated EIF2ALPHA (Ntd) and Protein Phosphatase 1A (D64A), Stabilized By G-Actin/Dnasei, PDB code: 9nb9:
In total only one binding site of Manganese was determined in the Viral Protein DP71L in Complex with Phosphorylated EIF2ALPHA (Ntd) and Protein Phosphatase 1A (D64A), Stabilized By G-Actin/Dnasei, PDB code: 9nb9:
Manganese binding site 1 out of 1 in 9nb9
Go back to
Manganese binding site 1 out
of 1 in the Viral Protein DP71L in Complex with Phosphorylated EIF2ALPHA (Ntd) and Protein Phosphatase 1A (D64A), Stabilized By G-Actin/Dnasei
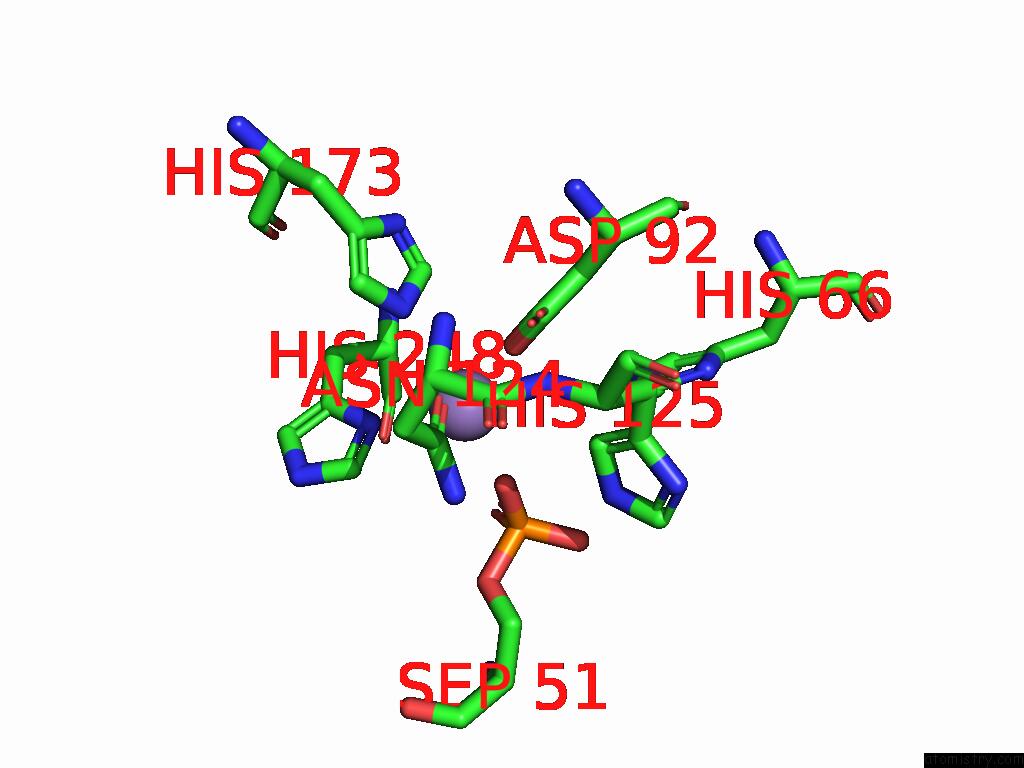
Mono view
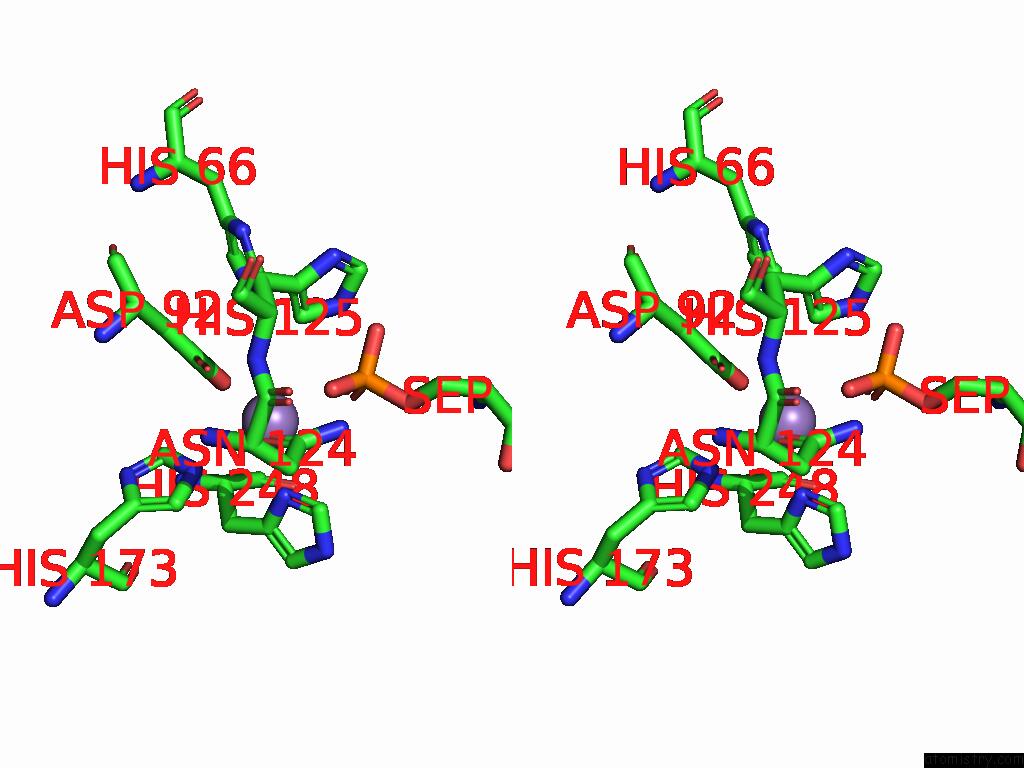
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Viral Protein DP71L in Complex with Phosphorylated EIF2ALPHA (Ntd) and Protein Phosphatase 1A (D64A), Stabilized By G-Actin/Dnasei within 5.0Å range:
|
Reference:
L.C.Reineke,
P.J.Zhu,
U.Dalwadi,
S.W.Dooling,
Y.Liu,
I.C.Wang,
S.Young-Baird,
J.Okoh,
S.K.Kuncha,
H.Zhou,
A.Kannan,
H.Park,
N.A.Debeaubien,
T.Croll,
D.J.Lee,
C.Arthur,
T.E.Dever,
P.Walter,
J.Chen,
A.Frost,
M.Costa-Mattioli.
Harnessing the Evolution of Proteostasis Networks to Reverse Cognitive Dysfunction. Biorxiv 2025.
ISSN: ISSN 2692-8205
PubMed: 40568171
DOI: 10.1101/2025.02.28.640897
Page generated: Sun Aug 17 02:48:46 2025
ISSN: ISSN 2692-8205
PubMed: 40568171
DOI: 10.1101/2025.02.28.640897
Last articles
Zn in 9QM9Zn in 9S44
Zn in 9OFE
Zn in 9OFC
Zn in 9OFD
Zn in 9OF1
Zn in 9OFB
Zn in 9N0J
Zn in 9M5X
Zn in 9LGI