Manganese »
PDB 8sln-8uwb »
8umc »
Manganese in PDB 8umc: Deinococcus Aerius TR0125 C-Glucosyl Deglycosidase (Cgd), Cryo-Em
Manganese Binding Sites:
The binding sites of Manganese atom in the Deinococcus Aerius TR0125 C-Glucosyl Deglycosidase (Cgd), Cryo-Em
(pdb code 8umc). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 2 binding sites of Manganese where determined in the Deinococcus Aerius TR0125 C-Glucosyl Deglycosidase (Cgd), Cryo-Em, PDB code: 8umc:
Jump to Manganese binding site number: 1; 2;
In total 2 binding sites of Manganese where determined in the Deinococcus Aerius TR0125 C-Glucosyl Deglycosidase (Cgd), Cryo-Em, PDB code: 8umc:
Jump to Manganese binding site number: 1; 2;
Manganese binding site 1 out of 2 in 8umc
Go back to
Manganese binding site 1 out
of 2 in the Deinococcus Aerius TR0125 C-Glucosyl Deglycosidase (Cgd), Cryo-Em
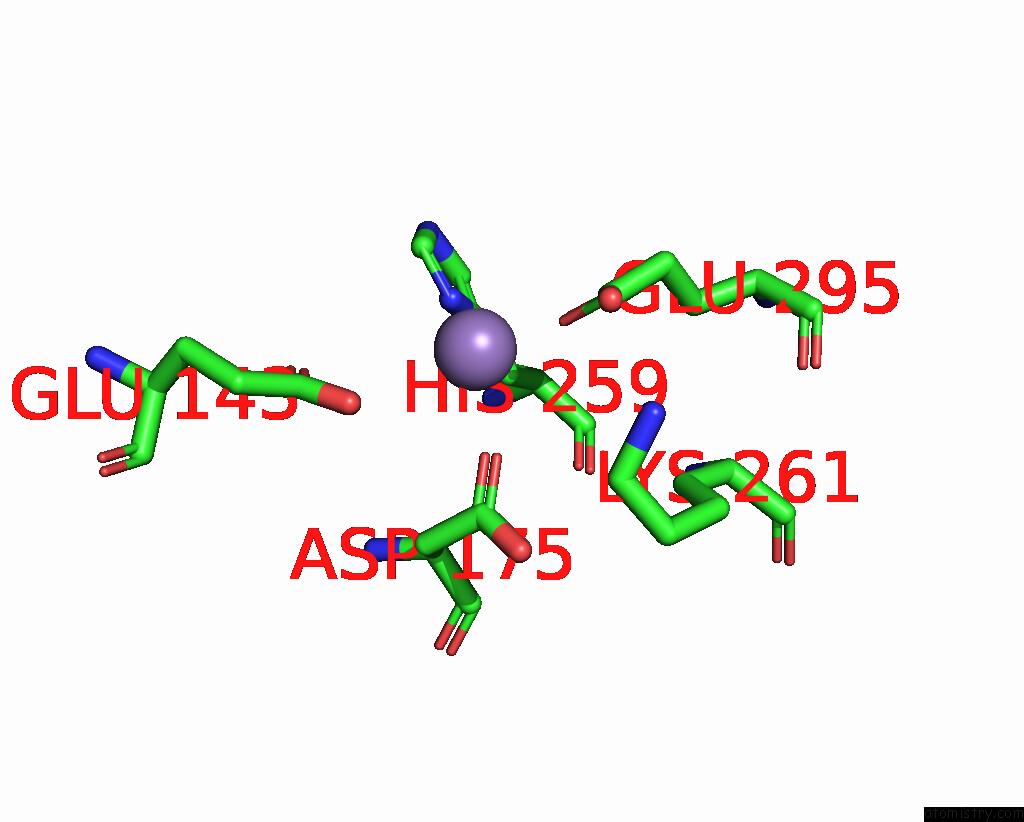
Mono view
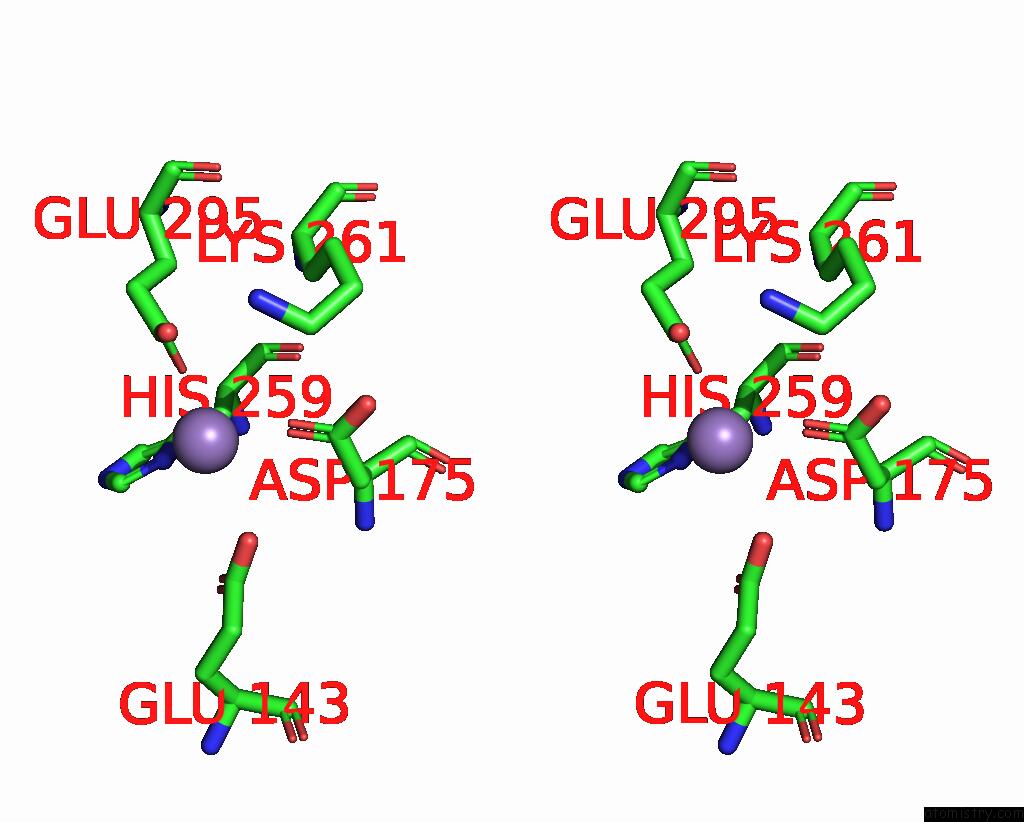
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Deinococcus Aerius TR0125 C-Glucosyl Deglycosidase (Cgd), Cryo-Em within 5.0Å range:
|
Manganese binding site 2 out of 2 in 8umc
Go back to
Manganese binding site 2 out
of 2 in the Deinococcus Aerius TR0125 C-Glucosyl Deglycosidase (Cgd), Cryo-Em
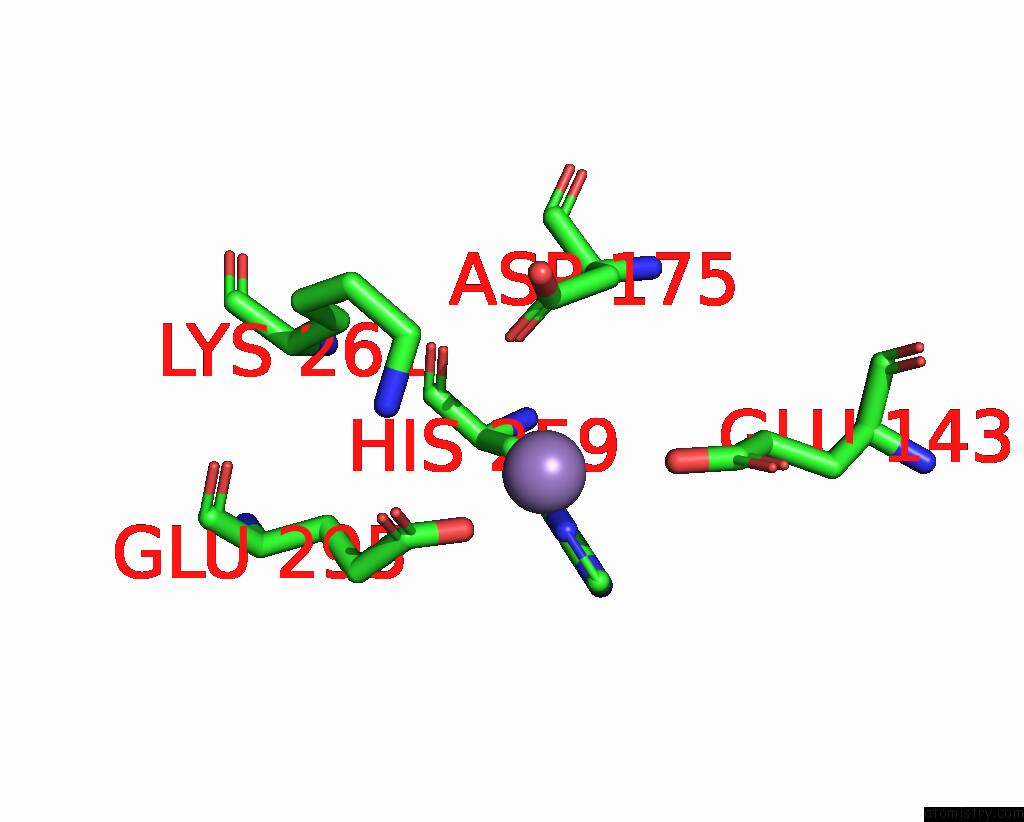
Mono view
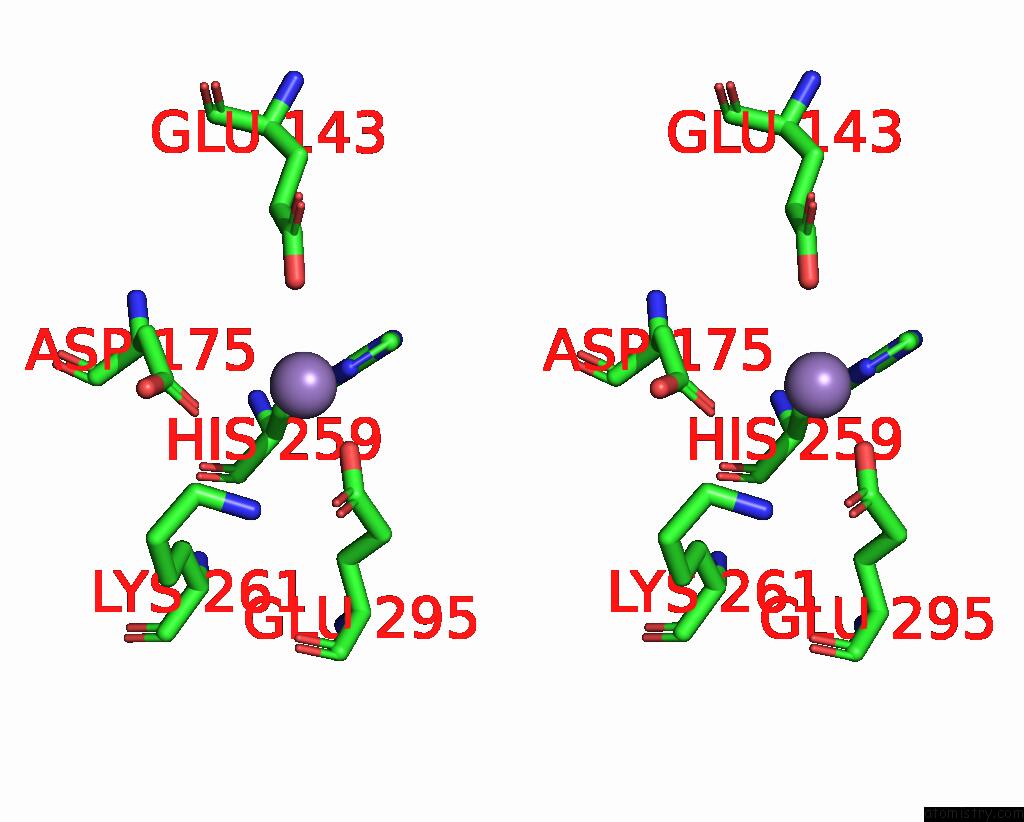
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Deinococcus Aerius TR0125 C-Glucosyl Deglycosidase (Cgd), Cryo-Em within 5.0Å range:
|
Reference:
V.Furlanetto,
D.C.Kalyani,
A.Kostelac,
J.Puc,
D.Haltrich,
B.M.Hallberg,
C.Divne.
Structural and Functional Characterization of A Gene Cluster Responsible For Deglycosylation of C-Glucosyl Flavonoids and Xanthonoids By Deinococcus Aerius. J.Mol.Biol. V. 436 68547 2024.
ISSN: ESSN 1089-8638
PubMed: 38508304
DOI: 10.1016/J.JMB.2024.168547
Page generated: Sun Oct 6 14:01:28 2024
ISSN: ESSN 1089-8638
PubMed: 38508304
DOI: 10.1016/J.JMB.2024.168547
Last articles
I in 4OA5I in 4OWA
I in 4P4V
I in 4OWC
I in 4P0D
I in 4OP3
I in 4OP2
I in 4OUC
I in 4OP1
I in 4OLH