Manganese »
PDB 8dl1-8f4c »
8e8d »
Manganese in PDB 8e8d: Human Dna Polymerase Eta-Dna-Ru-Ended Primer Ternary Mismatch Complex:Reaction with 10 Mm MN2+ For 60S
Enzymatic activity of Human Dna Polymerase Eta-Dna-Ru-Ended Primer Ternary Mismatch Complex:Reaction with 10 Mm MN2+ For 60S
All present enzymatic activity of Human Dna Polymerase Eta-Dna-Ru-Ended Primer Ternary Mismatch Complex:Reaction with 10 Mm MN2+ For 60S:
2.7.7.7;
2.7.7.7;
Protein crystallography data
The structure of Human Dna Polymerase Eta-Dna-Ru-Ended Primer Ternary Mismatch Complex:Reaction with 10 Mm MN2+ For 60S, PDB code: 8e8d
was solved by
C.Chang,
Y.Gao,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 31.43 / 2.09 |
| Space group | P 61 |
| Cell size a, b, c (Å), α, β, γ (°) | 98.48, 98.48, 81.68, 90, 90, 120 |
| R / Rfree (%) | 20 / 23.5 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Human Dna Polymerase Eta-Dna-Ru-Ended Primer Ternary Mismatch Complex:Reaction with 10 Mm MN2+ For 60S
(pdb code 8e8d). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 2 binding sites of Manganese where determined in the Human Dna Polymerase Eta-Dna-Ru-Ended Primer Ternary Mismatch Complex:Reaction with 10 Mm MN2+ For 60S, PDB code: 8e8d:
Jump to Manganese binding site number: 1; 2;
In total 2 binding sites of Manganese where determined in the Human Dna Polymerase Eta-Dna-Ru-Ended Primer Ternary Mismatch Complex:Reaction with 10 Mm MN2+ For 60S, PDB code: 8e8d:
Jump to Manganese binding site number: 1; 2;
Manganese binding site 1 out of 2 in 8e8d
Go back to
Manganese binding site 1 out
of 2 in the Human Dna Polymerase Eta-Dna-Ru-Ended Primer Ternary Mismatch Complex:Reaction with 10 Mm MN2+ For 60S
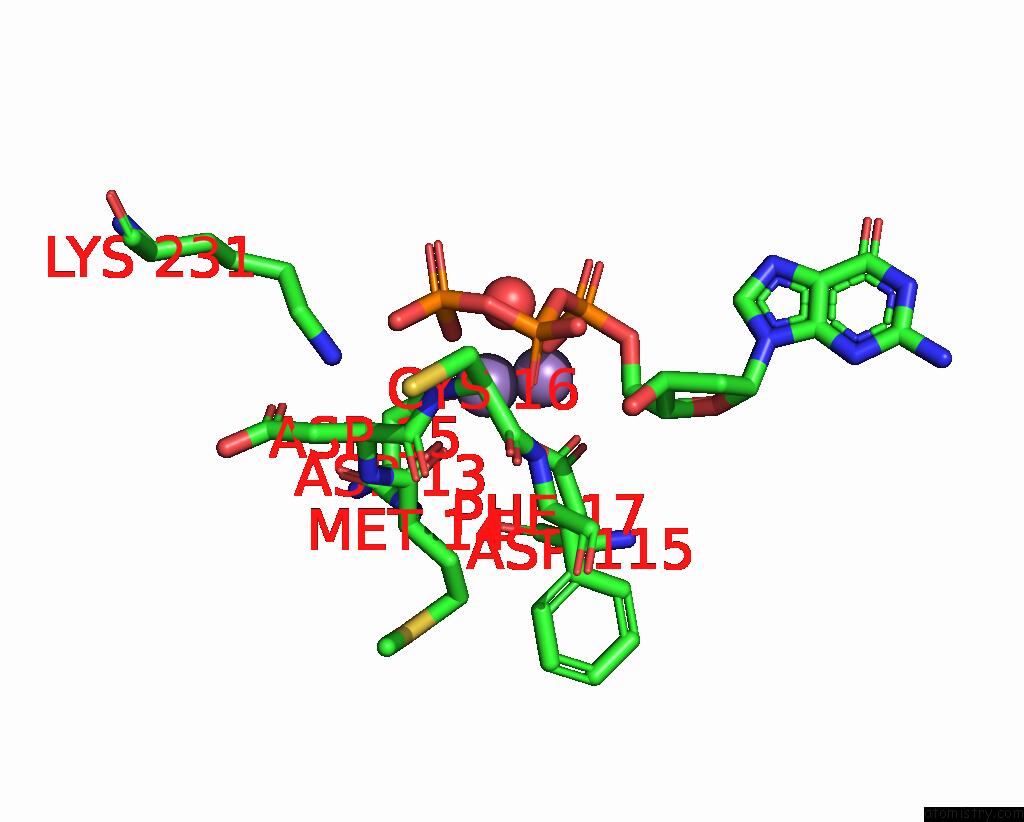
Mono view
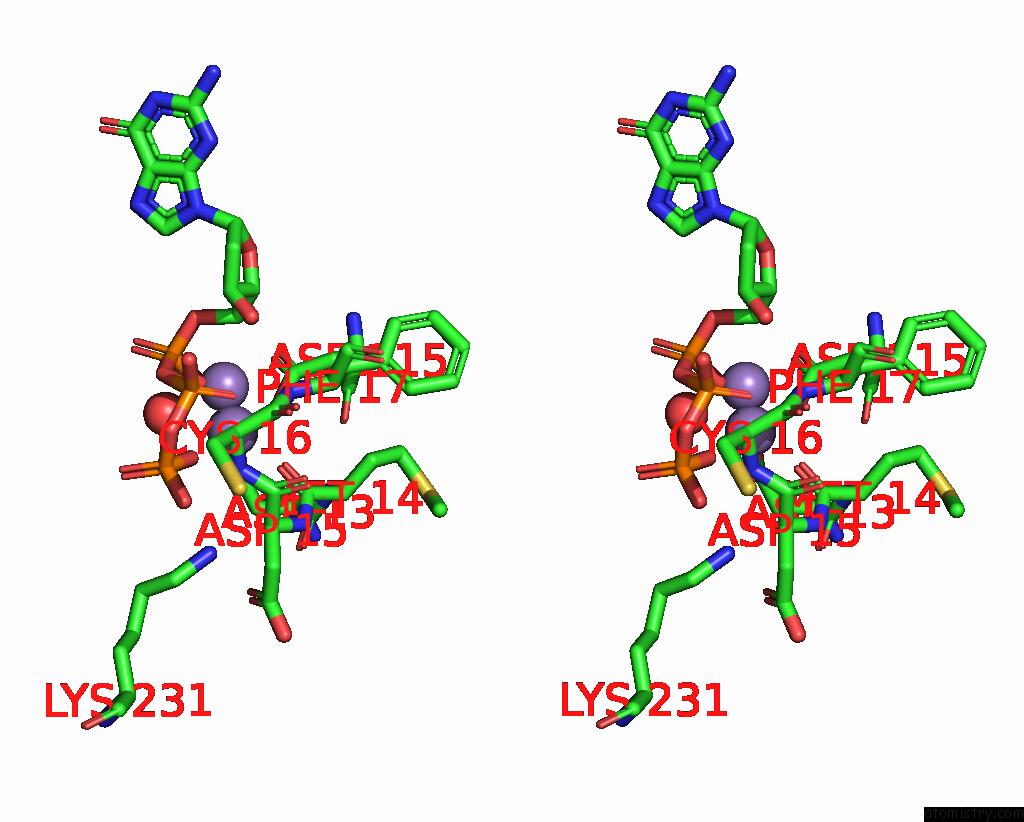
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Human Dna Polymerase Eta-Dna-Ru-Ended Primer Ternary Mismatch Complex:Reaction with 10 Mm MN2+ For 60S within 5.0Å range:
|
Manganese binding site 2 out of 2 in 8e8d
Go back to
Manganese binding site 2 out
of 2 in the Human Dna Polymerase Eta-Dna-Ru-Ended Primer Ternary Mismatch Complex:Reaction with 10 Mm MN2+ For 60S
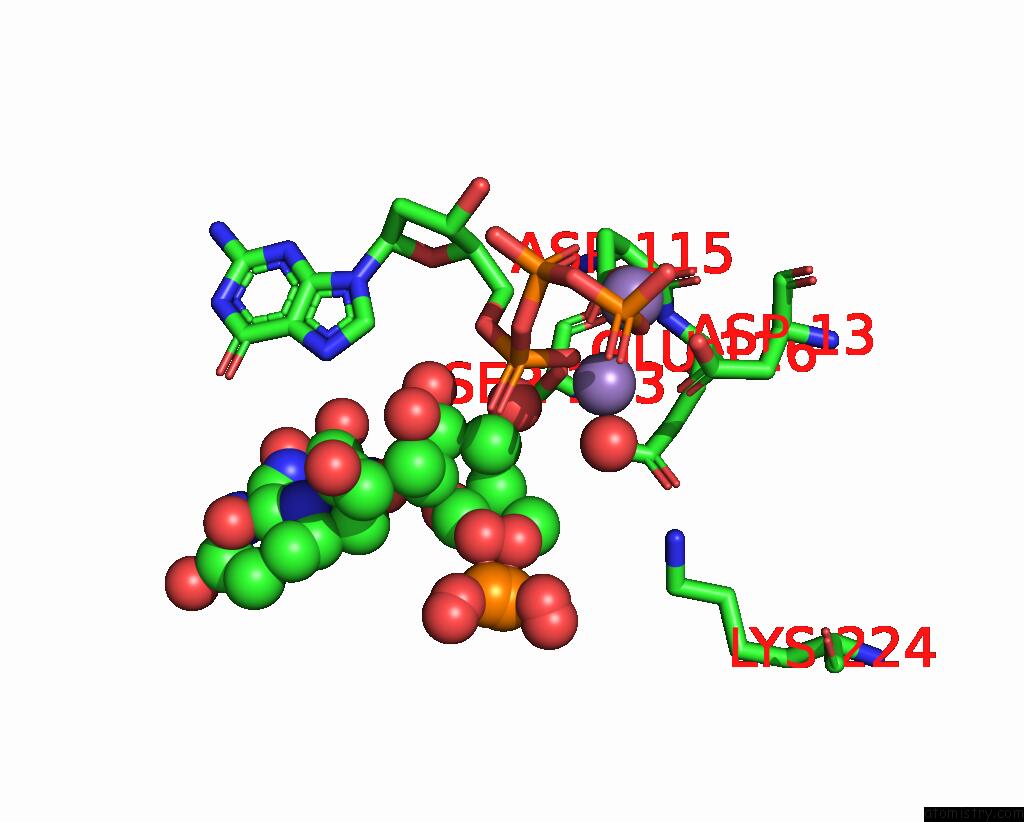
Mono view
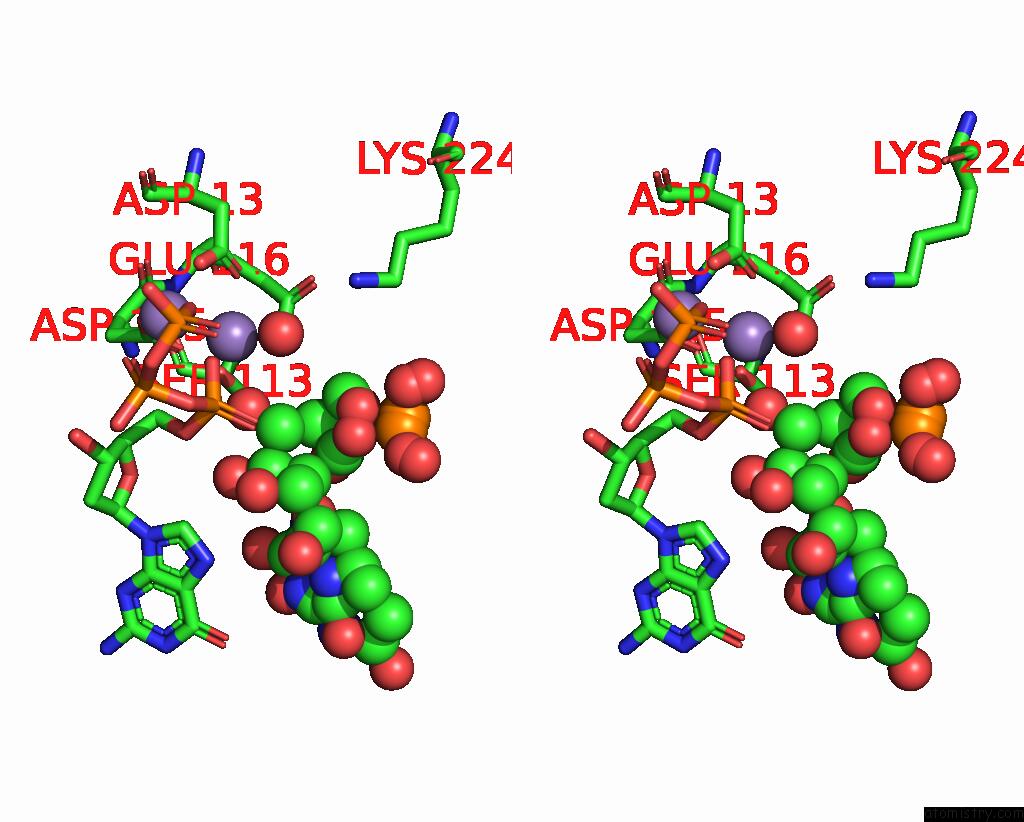
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Human Dna Polymerase Eta-Dna-Ru-Ended Primer Ternary Mismatch Complex:Reaction with 10 Mm MN2+ For 60S within 5.0Å range:
|
Reference:
C.Chang,
C.Lee Luo,
S.Eleraky,
A.Lin,
G.Zhou,
Y.Gao.
Primer Terminal Ribonucleotide Alters the Active Site Dynamics of Dna Polymerase Eta and Reduce Dna Synthesis Fidelity J.Biol.Chem. 02938 2023.
ISSN: ESSN 1083-351X
DOI: 10.1016/J.JBC.2023.102938
Page generated: Sun Oct 6 11:39:51 2024
ISSN: ESSN 1083-351X
DOI: 10.1016/J.JBC.2023.102938
Last articles
I in 4P9TI in 4PNX
I in 4PVQ
I in 4PVG
I in 4PGC
I in 4PNS
I in 4P4Z
I in 4P1E
I in 4P4X
I in 4P4Y