Manganese »
PDB 7z03-8awv »
7zz1 »
Manganese in PDB 7zz1: Cryo-Em Structure of "Ct React" Conformation of Lactococcus Lactis Pyruvate Carboxylase with Acetyl-Coa
Enzymatic activity of Cryo-Em Structure of "Ct React" Conformation of Lactococcus Lactis Pyruvate Carboxylase with Acetyl-Coa
All present enzymatic activity of Cryo-Em Structure of "Ct React" Conformation of Lactococcus Lactis Pyruvate Carboxylase with Acetyl-Coa:
6.4.1.1;
6.4.1.1;
Other elements in 7zz1:
The structure of Cryo-Em Structure of "Ct React" Conformation of Lactococcus Lactis Pyruvate Carboxylase with Acetyl-Coa also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Manganese Binding Sites:
The binding sites of Manganese atom in the Cryo-Em Structure of "Ct React" Conformation of Lactococcus Lactis Pyruvate Carboxylase with Acetyl-Coa
(pdb code 7zz1). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total only one binding site of Manganese was determined in the Cryo-Em Structure of "Ct React" Conformation of Lactococcus Lactis Pyruvate Carboxylase with Acetyl-Coa, PDB code: 7zz1:
In total only one binding site of Manganese was determined in the Cryo-Em Structure of "Ct React" Conformation of Lactococcus Lactis Pyruvate Carboxylase with Acetyl-Coa, PDB code: 7zz1:
Manganese binding site 1 out of 1 in 7zz1
Go back to
Manganese binding site 1 out
of 1 in the Cryo-Em Structure of "Ct React" Conformation of Lactococcus Lactis Pyruvate Carboxylase with Acetyl-Coa
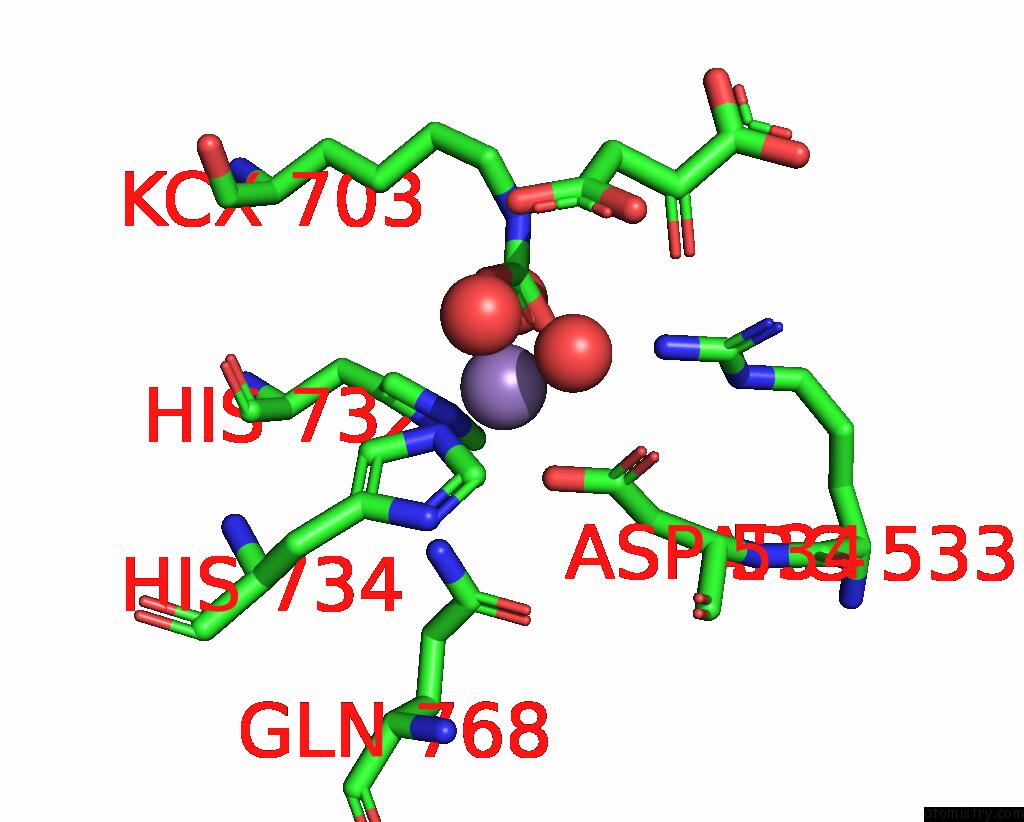
Mono view
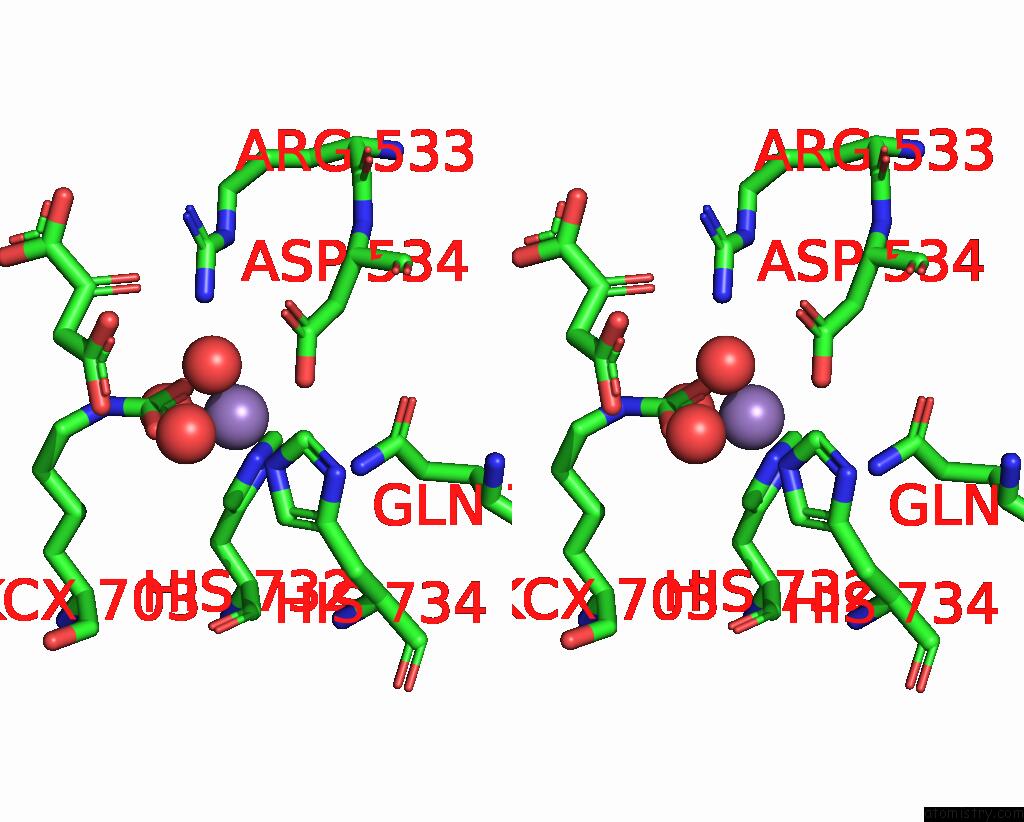
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Cryo-Em Structure of "Ct React" Conformation of Lactococcus Lactis Pyruvate Carboxylase with Acetyl-Coa within 5.0Å range:
|
Reference:
J.P.Lopez-Alonso,
M.Lazaro,
D.Gil-Carton,
P.H.Choi,
L.Tong,
M.Valle.
Cryoem Structural Exploration of Catalytically Active Enzyme Pyruvate Carboxylase. Nat Commun V. 13 6185 2022.
ISSN: ESSN 2041-1723
PubMed: 36261450
DOI: 10.1038/S41467-022-33987-2
Page generated: Sun Oct 6 11:15:02 2024
ISSN: ESSN 2041-1723
PubMed: 36261450
DOI: 10.1038/S41467-022-33987-2
Last articles
K in 7QQPK in 7QQO
K in 7QK5
K in 7QIX
K in 7QNO
K in 7QIY
K in 7Q3X
K in 7QDN
K in 7QF6
K in 7Q0G