Manganese »
PDB 7x9j-7yzp »
7yzi »
Manganese in PDB 7yzi: Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya
Enzymatic activity of Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya
All present enzymatic activity of Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya:
4.6.1.1;
4.6.1.1;
Manganese Binding Sites:
The binding sites of Manganese atom in the Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya
(pdb code 7yzi). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 4 binding sites of Manganese where determined in the Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya, PDB code: 7yzi:
Jump to Manganese binding site number: 1; 2; 3; 4;
In total 4 binding sites of Manganese where determined in the Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya, PDB code: 7yzi:
Jump to Manganese binding site number: 1; 2; 3; 4;
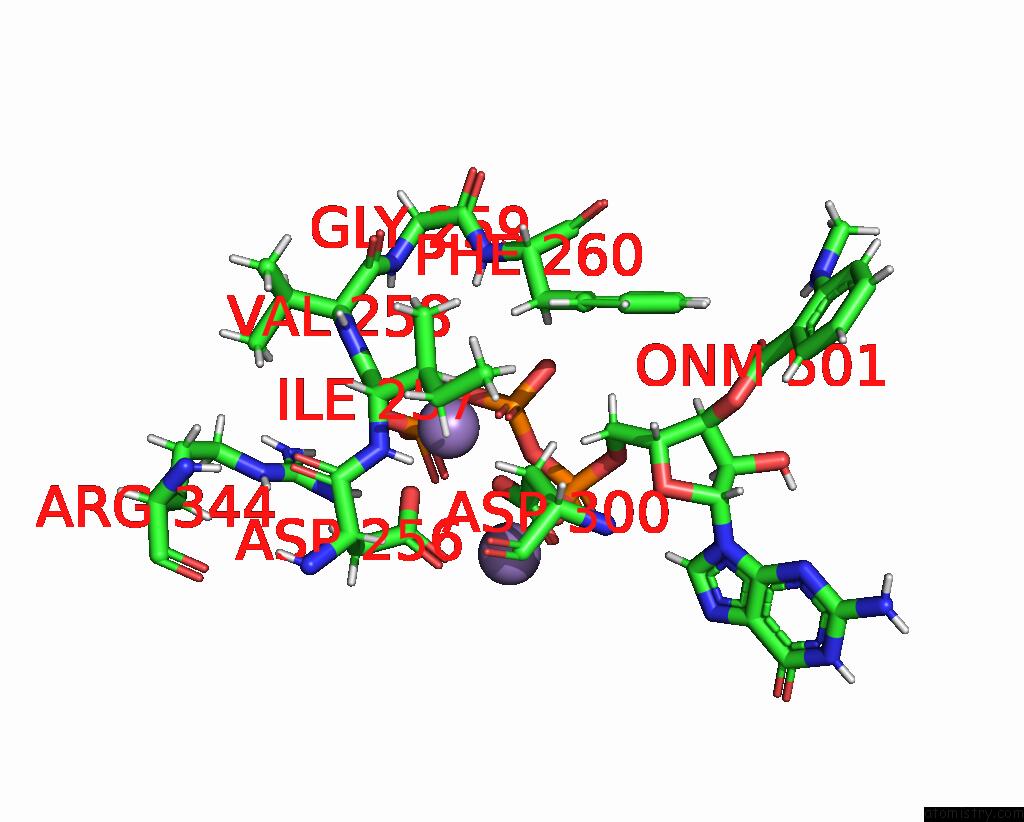
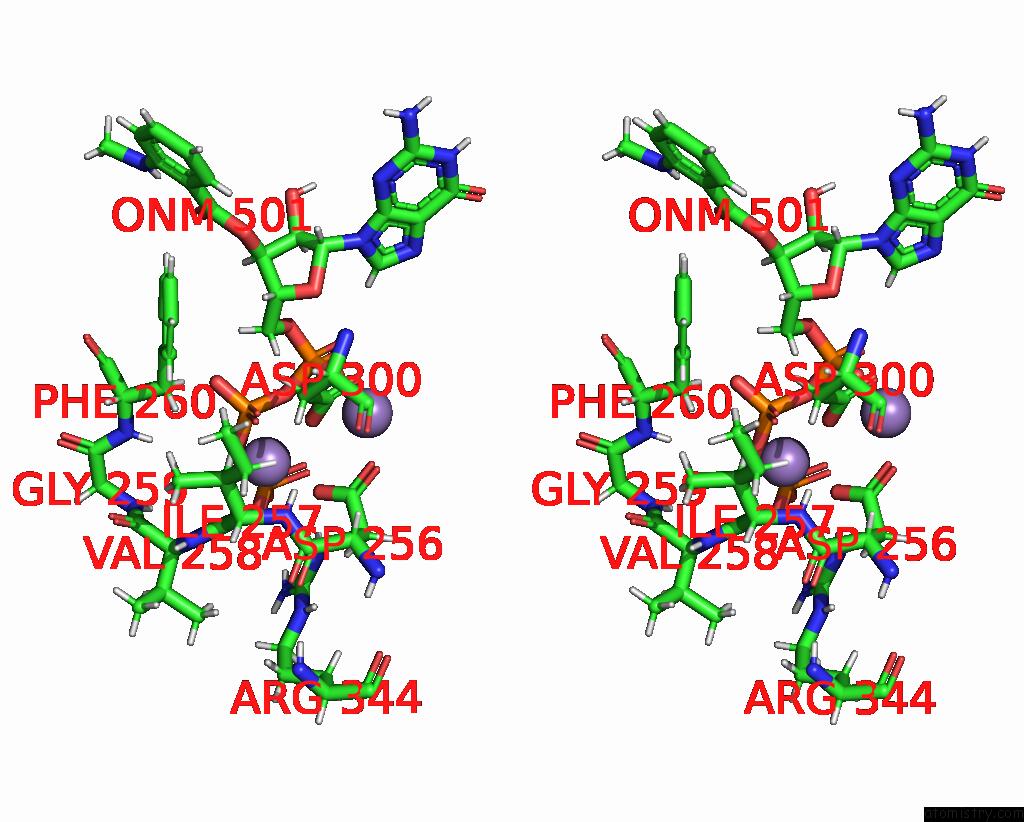
Manganese binding site 1 out of 4 in 7yzi
Go back to
Manganese binding site 1 out
of 4 in the Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya within 5.0Å range:
|
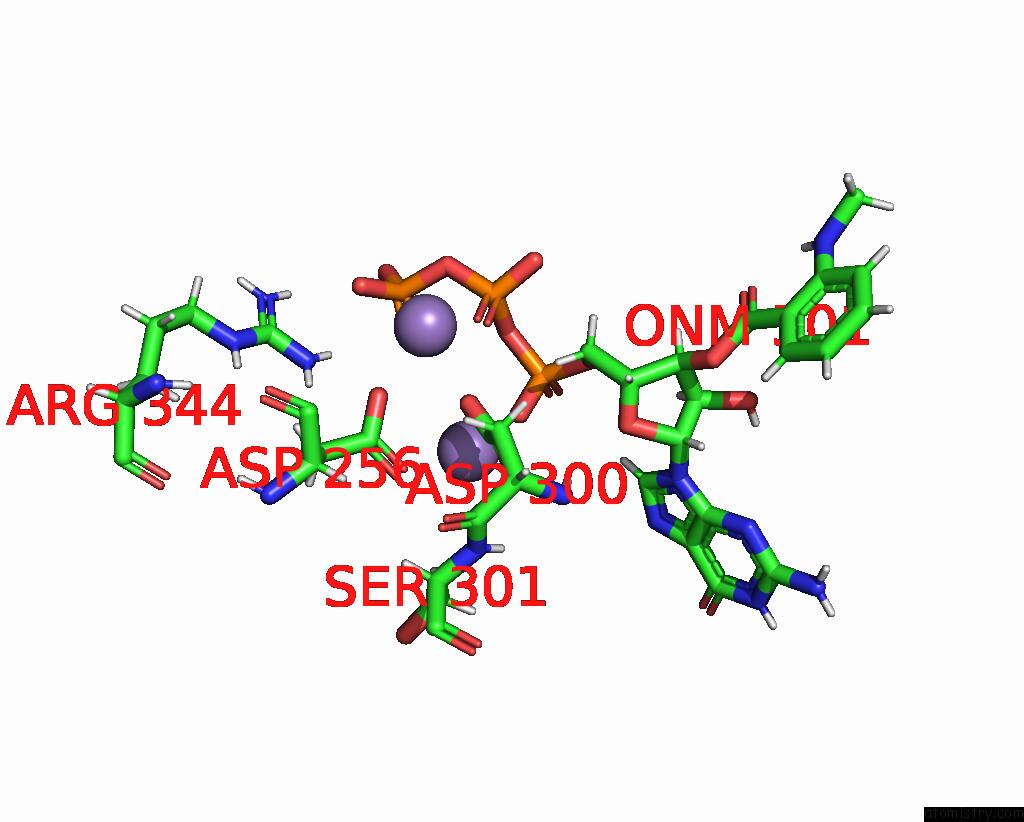
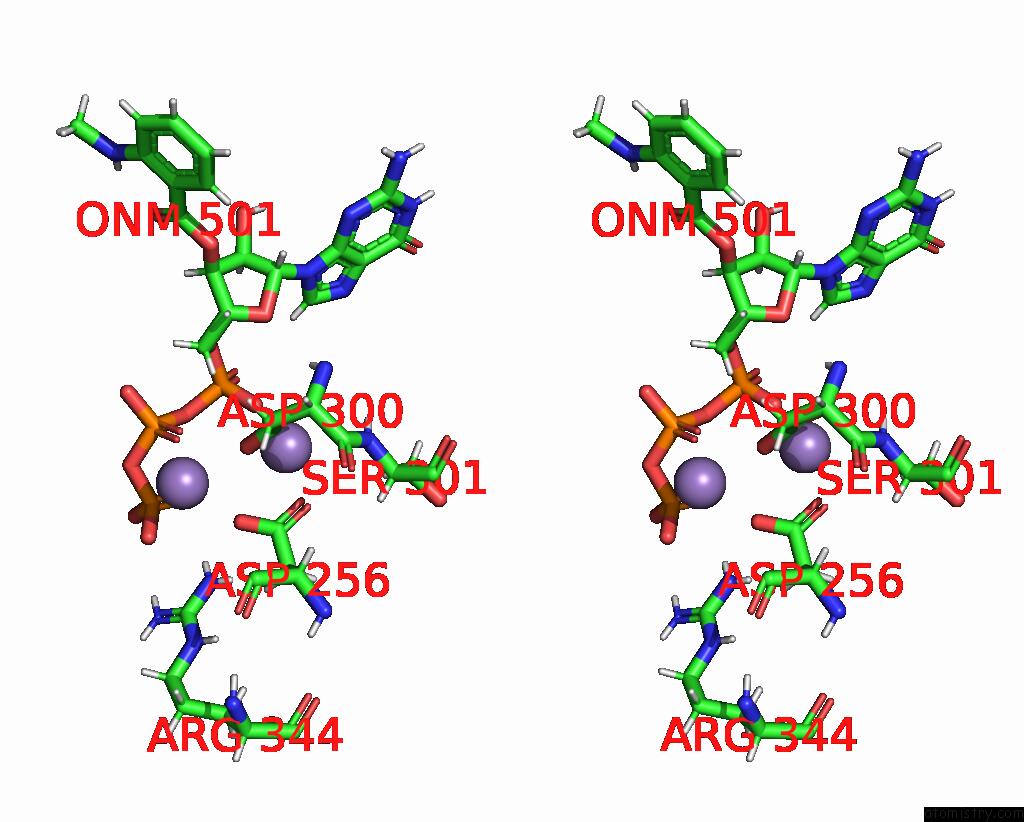
Manganese binding site 2 out of 4 in 7yzi
Go back to
Manganese binding site 2 out
of 4 in the Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya within 5.0Å range:
|
Manganese binding site 3 out of 4 in 7yzi
Go back to
Manganese binding site 3 out
of 4 in the Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya
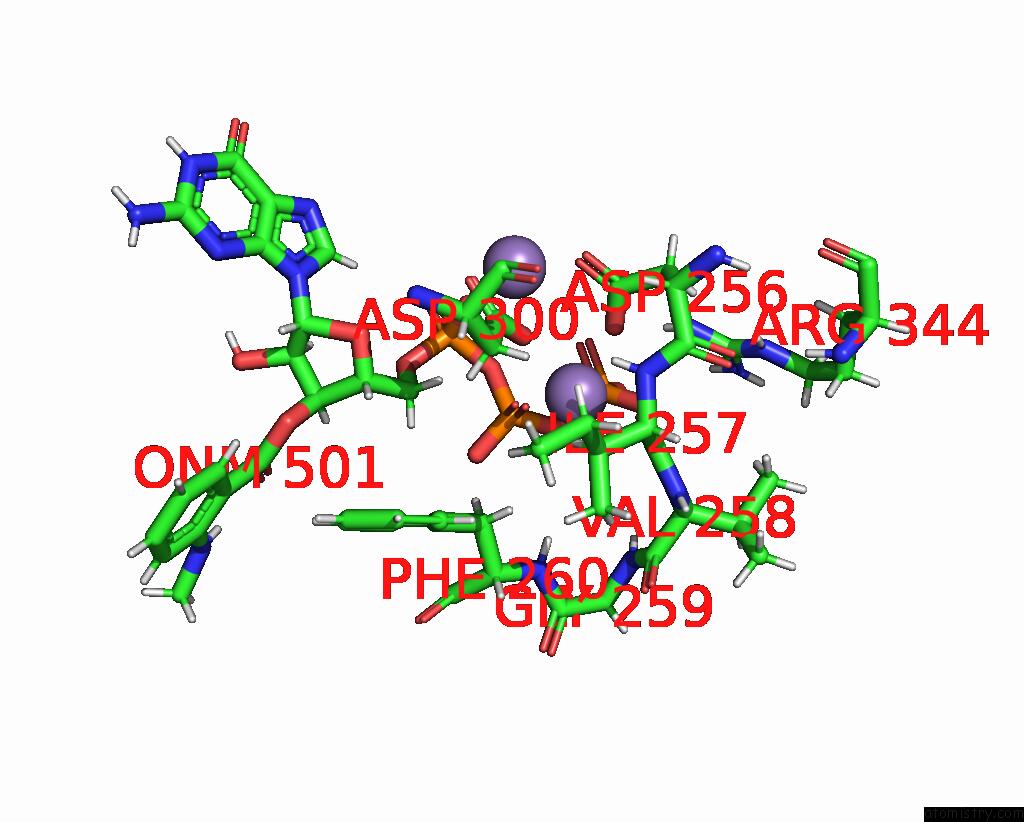
Mono view
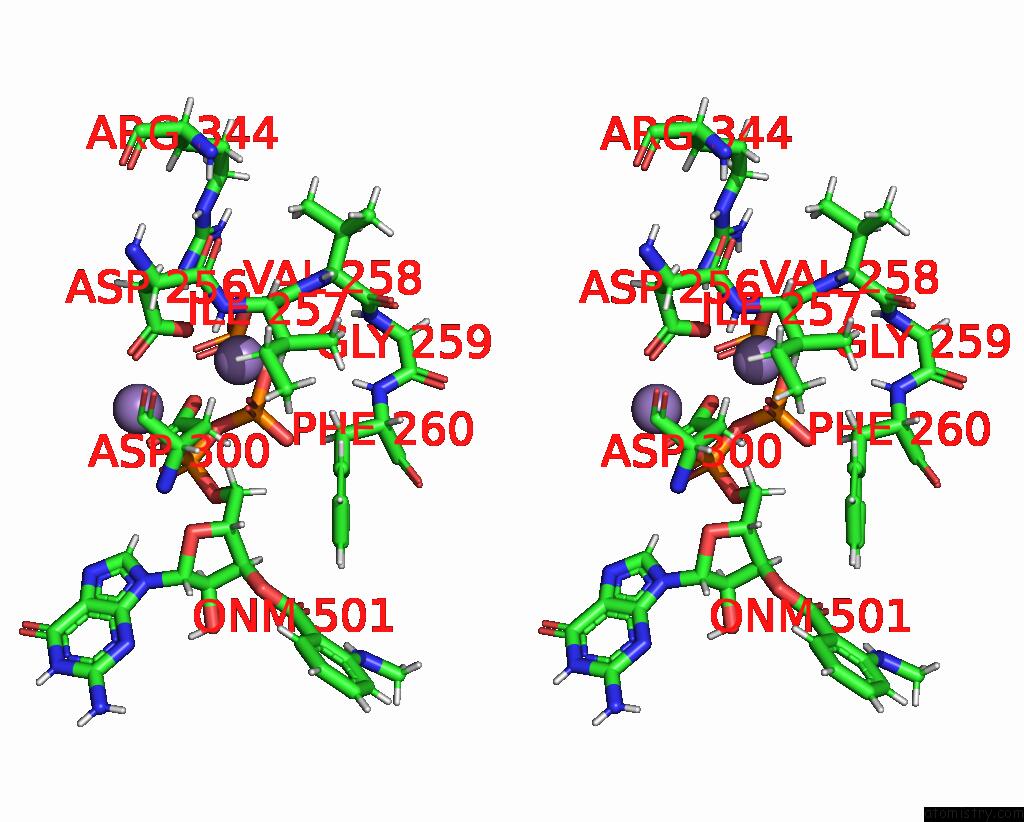
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya within 5.0Å range:
|
Manganese binding site 4 out of 4 in 7yzi
Go back to
Manganese binding site 4 out
of 4 in the Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya
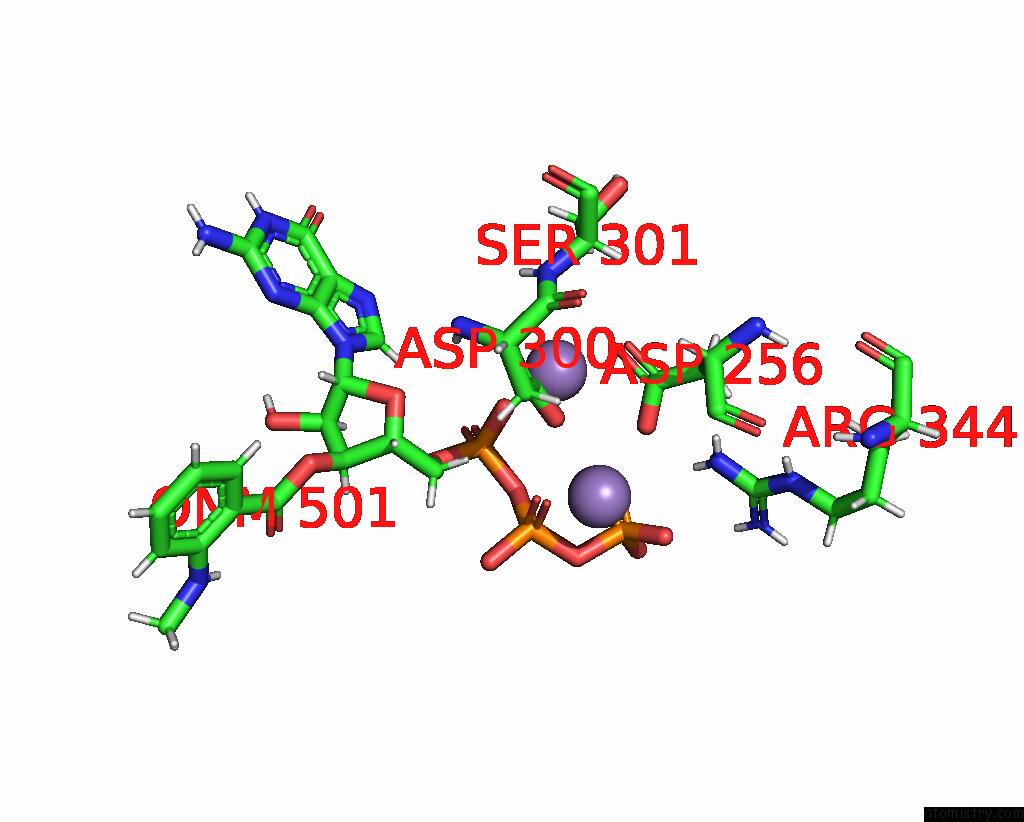
Mono view
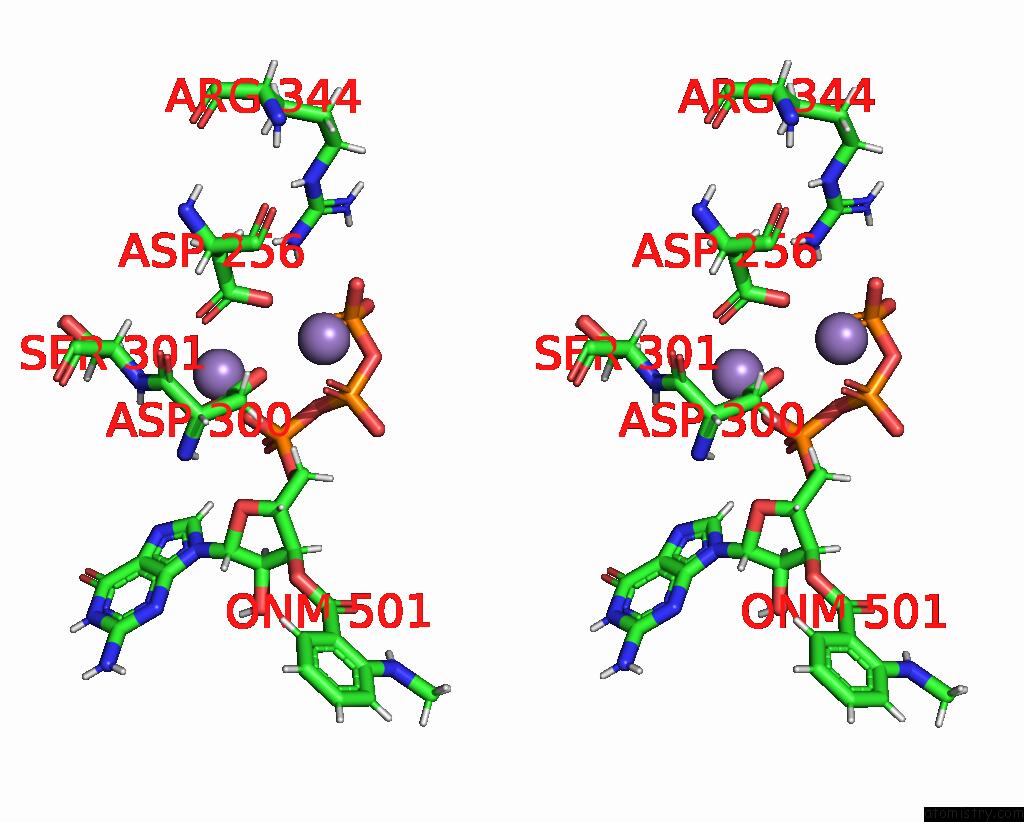
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya within 5.0Å range:
|
Reference:
V.Mehta,
B.Khanppnavar,
D.Schuster,
I.Kantarci,
I.Vercellino,
A.Kosturanova,
T.Iype,
S.Stefanic,
P.Picotti,
V.M.Korkhov.
Structure of Mycobacterium Tuberculosis Cya, An Evolutionary Ancestor of the Mammalian Membrane Adenylyl Cyclases. Elife V. 11 2022.
ISSN: ESSN 2050-084X
PubMed: 35980026
DOI: 10.7554/ELIFE.77032
Page generated: Sun Aug 17 00:03:36 2025
ISSN: ESSN 2050-084X
PubMed: 35980026
DOI: 10.7554/ELIFE.77032
Last articles
Ni in 5XLFNi in 5XLG
Ni in 5XLE
Ni in 5XGZ
Ni in 5XF9
Ni in 5XFA
Ni in 5XGF
Ni in 5XEW
Ni in 5X7R
Ni in 5X2U