Manganese »
PDB 7x9j-7yzp »
7xgc »
Manganese in PDB 7xgc: Crystal Structure of Influenza Polymerase Acidic Subunit N-Terminal Domain Crystallized By Ammonium Sulfate with Glycan
Protein crystallography data
The structure of Crystal Structure of Influenza Polymerase Acidic Subunit N-Terminal Domain Crystallized By Ammonium Sulfate with Glycan, PDB code: 7xgc
was solved by
Y.Guo,
T.Hoshino,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 37.54 / 1.60 |
| Space group | P 41 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 66.177, 66.177, 125.727, 90, 90, 90 |
| R / Rfree (%) | 20.5 / 22.8 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Crystal Structure of Influenza Polymerase Acidic Subunit N-Terminal Domain Crystallized By Ammonium Sulfate with Glycan
(pdb code 7xgc). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 2 binding sites of Manganese where determined in the Crystal Structure of Influenza Polymerase Acidic Subunit N-Terminal Domain Crystallized By Ammonium Sulfate with Glycan, PDB code: 7xgc:
Jump to Manganese binding site number: 1; 2;
In total 2 binding sites of Manganese where determined in the Crystal Structure of Influenza Polymerase Acidic Subunit N-Terminal Domain Crystallized By Ammonium Sulfate with Glycan, PDB code: 7xgc:
Jump to Manganese binding site number: 1; 2;
Manganese binding site 1 out of 2 in 7xgc
Go back to
Manganese binding site 1 out
of 2 in the Crystal Structure of Influenza Polymerase Acidic Subunit N-Terminal Domain Crystallized By Ammonium Sulfate with Glycan
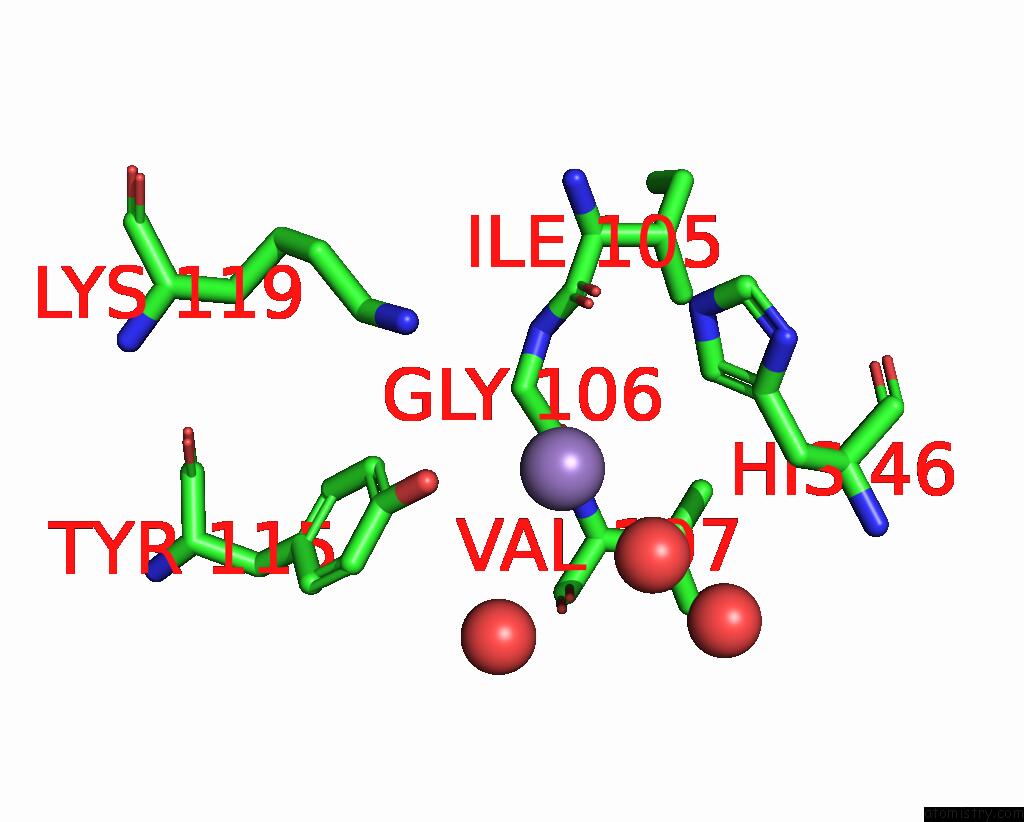
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Crystal Structure of Influenza Polymerase Acidic Subunit N-Terminal Domain Crystallized By Ammonium Sulfate with Glycan within 5.0Å range:
|
Manganese binding site 2 out of 2 in 7xgc
Go back to
Manganese binding site 2 out
of 2 in the Crystal Structure of Influenza Polymerase Acidic Subunit N-Terminal Domain Crystallized By Ammonium Sulfate with Glycan

Mono view
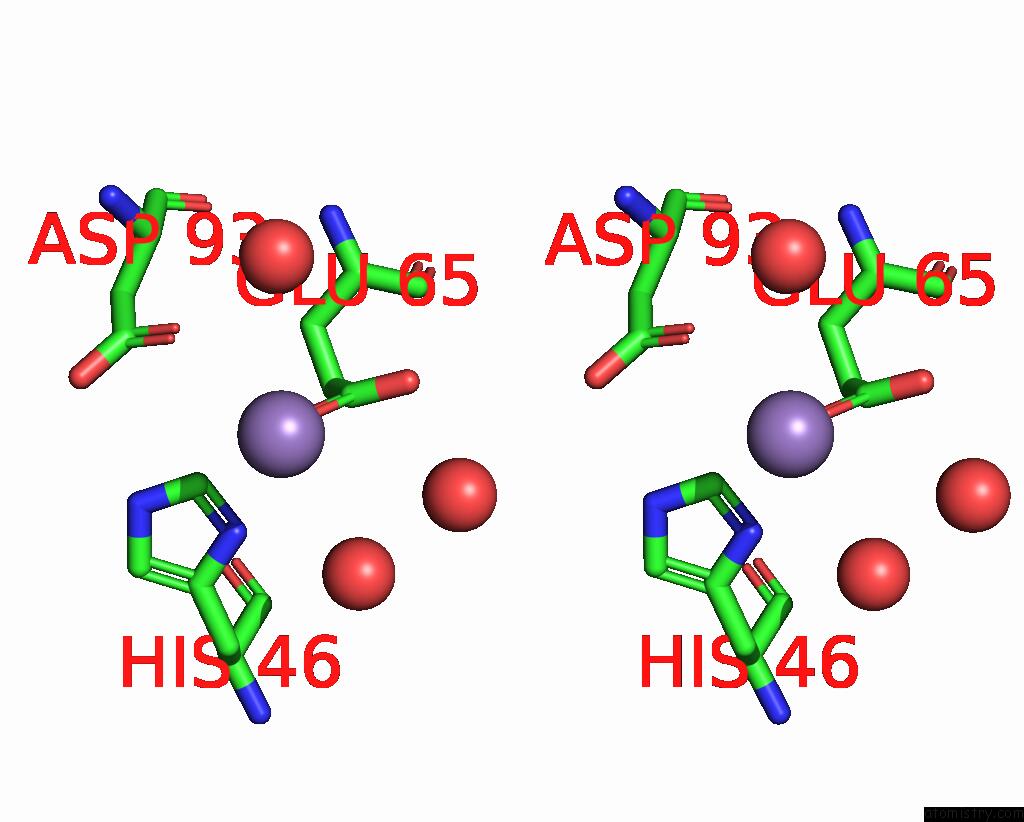
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Crystal Structure of Influenza Polymerase Acidic Subunit N-Terminal Domain Crystallized By Ammonium Sulfate with Glycan within 5.0Å range:
|
Reference:
Y.Guo,
T.Hoshino.
Influence of Glycan Agents on Protein Crystallization with Ammonium Sulfate Cryst.Growth Des. 2022.
ISSN: ESSN 1528-7505
DOI: 10.1021/ACS.CGD.2C00901
Page generated: Sat Aug 16 23:59:03 2025
ISSN: ESSN 1528-7505
DOI: 10.1021/ACS.CGD.2C00901
Last articles
Ni in 5XGFNi in 5XEW
Ni in 5X7R
Ni in 5X2U
Ni in 5X7S
Ni in 5VVL
Ni in 5X7Q
Ni in 5X7P
Ni in 5X7O
Ni in 5X57