Manganese »
PDB 6zbn-7bm0 »
7aii »
Manganese in PDB 7aii: Hiv-1 Reverse Transcriptase Complex with Dna and L-Methionine Tenofovir with Bound Manganese
Enzymatic activity of Hiv-1 Reverse Transcriptase Complex with Dna and L-Methionine Tenofovir with Bound Manganese
All present enzymatic activity of Hiv-1 Reverse Transcriptase Complex with Dna and L-Methionine Tenofovir with Bound Manganese:
2.7.7.49; 2.7.7.7; 3.1.13.2; 3.1.26.13; 3.4.23.16;
2.7.7.49; 2.7.7.7; 3.1.13.2; 3.1.26.13; 3.4.23.16;
Protein crystallography data
The structure of Hiv-1 Reverse Transcriptase Complex with Dna and L-Methionine Tenofovir with Bound Manganese, PDB code: 7aii
was solved by
W.Gu,
S.E.Martinez,
H.Nguyen,
H.Xu,
P.Herdewijn,
S.De Jonghe,
K.Das,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 69.78 / 2.62 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 89.48, 132.67, 139.02, 90, 98.62, 90 |
| R / Rfree (%) | 22.3 / 25.5 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Hiv-1 Reverse Transcriptase Complex with Dna and L-Methionine Tenofovir with Bound Manganese
(pdb code 7aii). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 4 binding sites of Manganese where determined in the Hiv-1 Reverse Transcriptase Complex with Dna and L-Methionine Tenofovir with Bound Manganese, PDB code: 7aii:
Jump to Manganese binding site number: 1; 2; 3; 4;
In total 4 binding sites of Manganese where determined in the Hiv-1 Reverse Transcriptase Complex with Dna and L-Methionine Tenofovir with Bound Manganese, PDB code: 7aii:
Jump to Manganese binding site number: 1; 2; 3; 4;
Manganese binding site 1 out of 4 in 7aii
Go back to
Manganese binding site 1 out
of 4 in the Hiv-1 Reverse Transcriptase Complex with Dna and L-Methionine Tenofovir with Bound Manganese
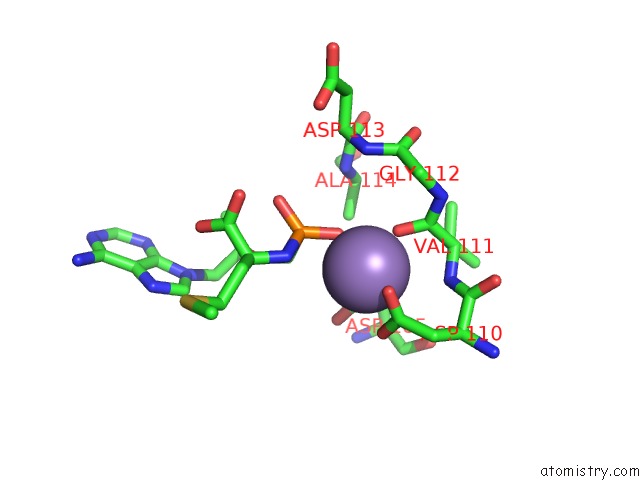
Mono view
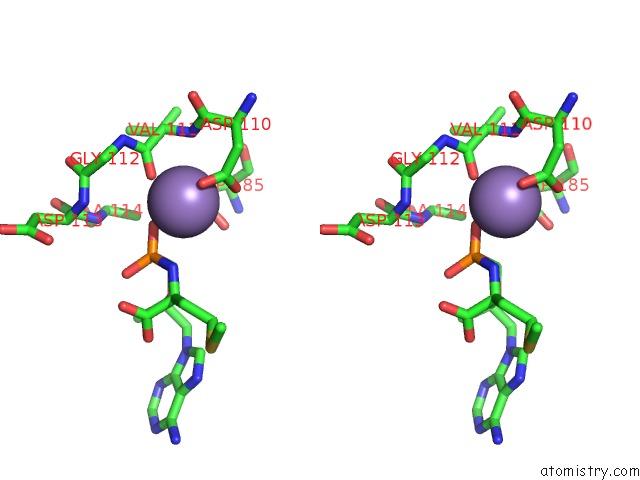
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Hiv-1 Reverse Transcriptase Complex with Dna and L-Methionine Tenofovir with Bound Manganese within 5.0Å range:
|
Manganese binding site 2 out of 4 in 7aii
Go back to
Manganese binding site 2 out
of 4 in the Hiv-1 Reverse Transcriptase Complex with Dna and L-Methionine Tenofovir with Bound Manganese
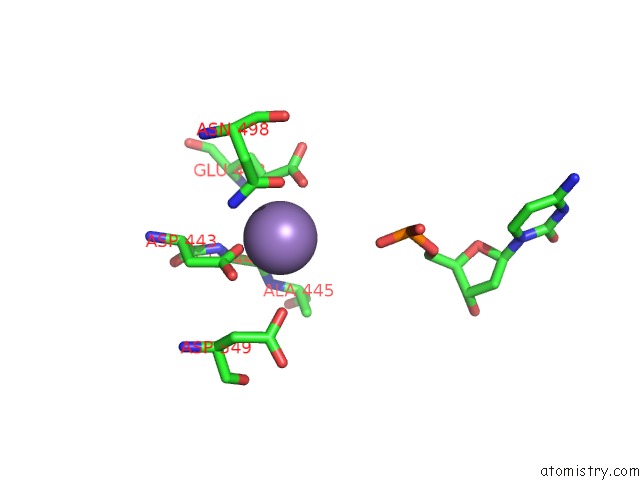
Mono view
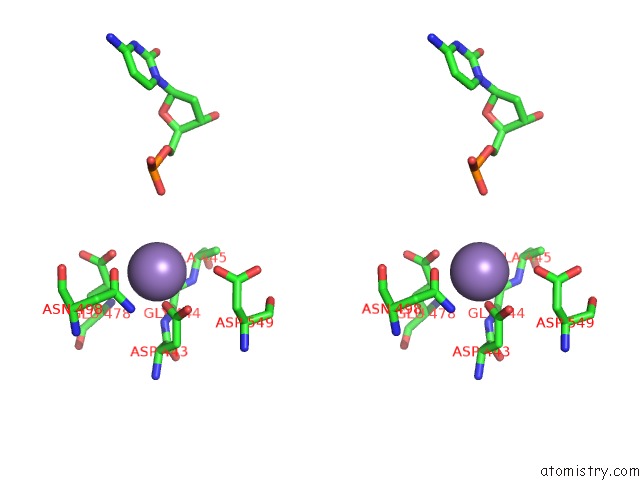
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Hiv-1 Reverse Transcriptase Complex with Dna and L-Methionine Tenofovir with Bound Manganese within 5.0Å range:
|
Manganese binding site 3 out of 4 in 7aii
Go back to
Manganese binding site 3 out
of 4 in the Hiv-1 Reverse Transcriptase Complex with Dna and L-Methionine Tenofovir with Bound Manganese
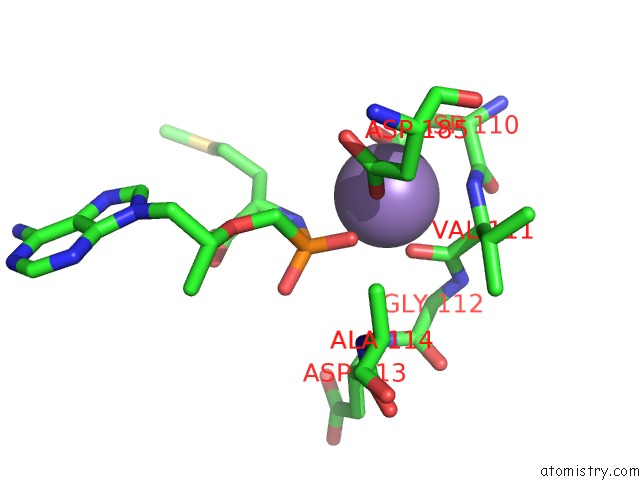
Mono view
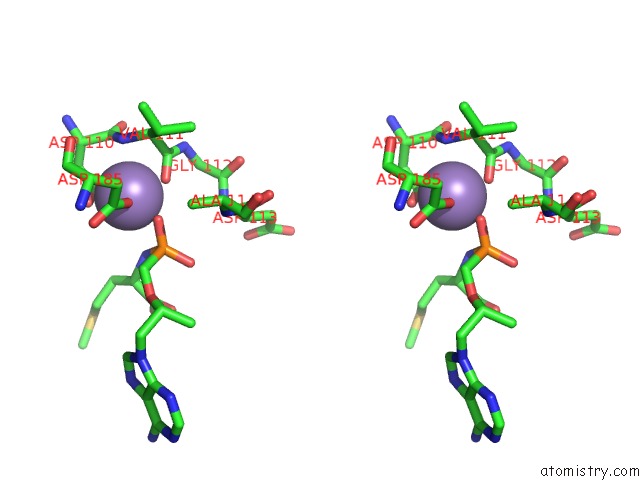
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Hiv-1 Reverse Transcriptase Complex with Dna and L-Methionine Tenofovir with Bound Manganese within 5.0Å range:
|
Manganese binding site 4 out of 4 in 7aii
Go back to
Manganese binding site 4 out
of 4 in the Hiv-1 Reverse Transcriptase Complex with Dna and L-Methionine Tenofovir with Bound Manganese
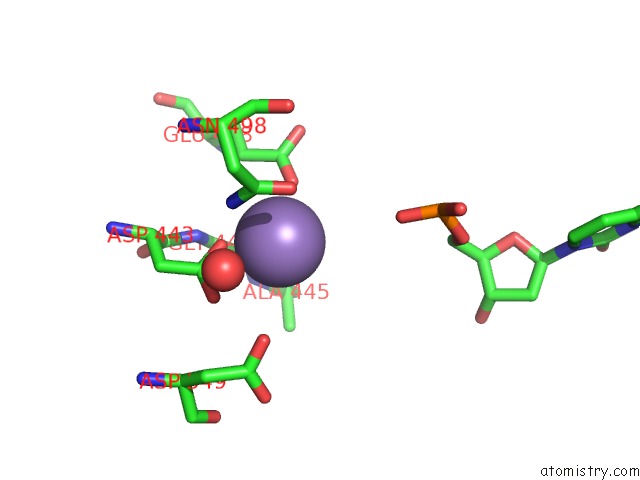
Mono view
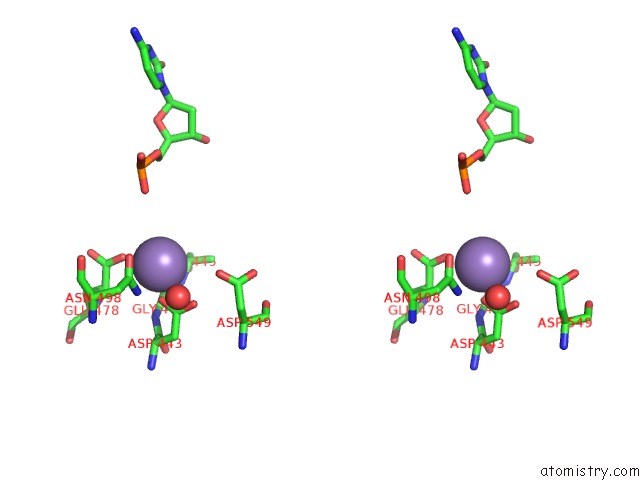
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of Hiv-1 Reverse Transcriptase Complex with Dna and L-Methionine Tenofovir with Bound Manganese within 5.0Å range:
|
Reference:
W.Gu,
S.Martinez,
H.Nguyen,
H.Xu,
P.Herdewijn,
S.De Jonghe,
K.Das.
Tenofovir-Amino Acid Conjugates Act As Polymerase Substrates-Implications For Avoiding Cellular Phosphorylation in the Discovery of Nucleotide Analogues. J.Med.Chem. 2020.
ISSN: ISSN 0022-2623
PubMed: 33356231
DOI: 10.1021/ACS.JMEDCHEM.0C01747
Page generated: Sun Oct 6 08:04:01 2024
ISSN: ISSN 0022-2623
PubMed: 33356231
DOI: 10.1021/ACS.JMEDCHEM.0C01747
Last articles
K in 6HTGK in 6HTK
K in 6HT8
K in 6HSZ
K in 6HSH
K in 6HSK
K in 6HSG
K in 6HSF
K in 6HRQ
K in 6HQY