Manganese »
PDB 6tzp-6vf4 »
6vaw »
Manganese in PDB 6vaw: Peanut Lectin Complexed with N-Beta-D-Galactopyranosyl-L-Succinamoyl Derivative (Ngs)
Protein crystallography data
The structure of Peanut Lectin Complexed with N-Beta-D-Galactopyranosyl-L-Succinamoyl Derivative (Ngs), PDB code: 6vaw
was solved by
L.H.Otero,
E.D.Primo,
A.J.Cagnoni,
S.Klinke,
F.A.Goldbaum,
M.L.Uhrig,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.42 / 1.75 |
| Space group | P 2 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 76.222, 125.052, 126.844, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.2 / 23.2 |
Other elements in 6vaw:
The structure of Peanut Lectin Complexed with N-Beta-D-Galactopyranosyl-L-Succinamoyl Derivative (Ngs) also contains other interesting chemical elements:
| Calcium | (Ca) | 4 atoms |
Manganese Binding Sites:
The binding sites of Manganese atom in the Peanut Lectin Complexed with N-Beta-D-Galactopyranosyl-L-Succinamoyl Derivative (Ngs)
(pdb code 6vaw). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 4 binding sites of Manganese where determined in the Peanut Lectin Complexed with N-Beta-D-Galactopyranosyl-L-Succinamoyl Derivative (Ngs), PDB code: 6vaw:
Jump to Manganese binding site number: 1; 2; 3; 4;
In total 4 binding sites of Manganese where determined in the Peanut Lectin Complexed with N-Beta-D-Galactopyranosyl-L-Succinamoyl Derivative (Ngs), PDB code: 6vaw:
Jump to Manganese binding site number: 1; 2; 3; 4;
Manganese binding site 1 out of 4 in 6vaw
Go back to
Manganese binding site 1 out
of 4 in the Peanut Lectin Complexed with N-Beta-D-Galactopyranosyl-L-Succinamoyl Derivative (Ngs)
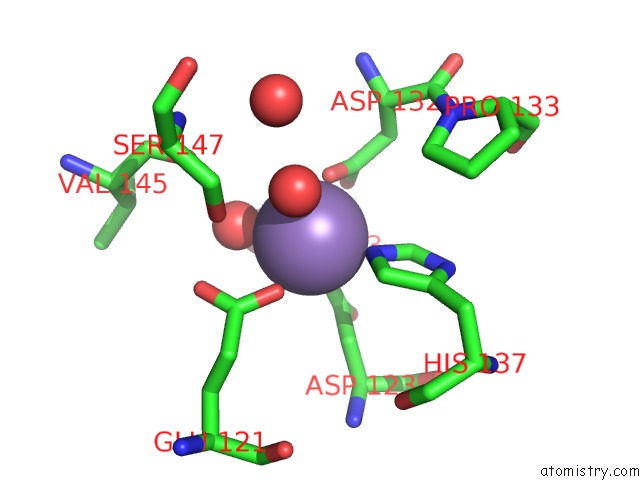
Mono view
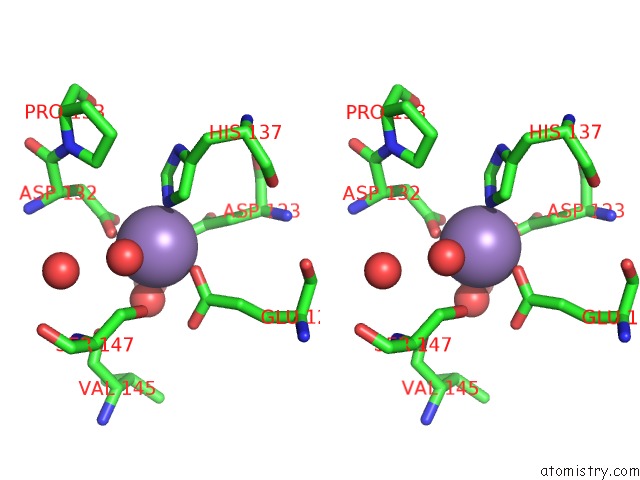
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Peanut Lectin Complexed with N-Beta-D-Galactopyranosyl-L-Succinamoyl Derivative (Ngs) within 5.0Å range:
|
Manganese binding site 2 out of 4 in 6vaw
Go back to
Manganese binding site 2 out
of 4 in the Peanut Lectin Complexed with N-Beta-D-Galactopyranosyl-L-Succinamoyl Derivative (Ngs)
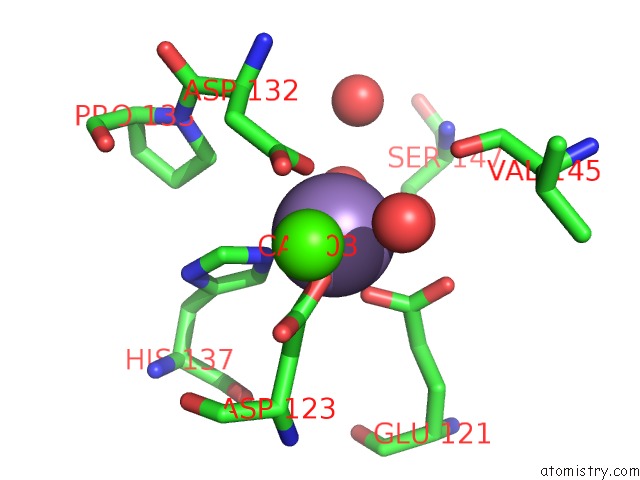
Mono view
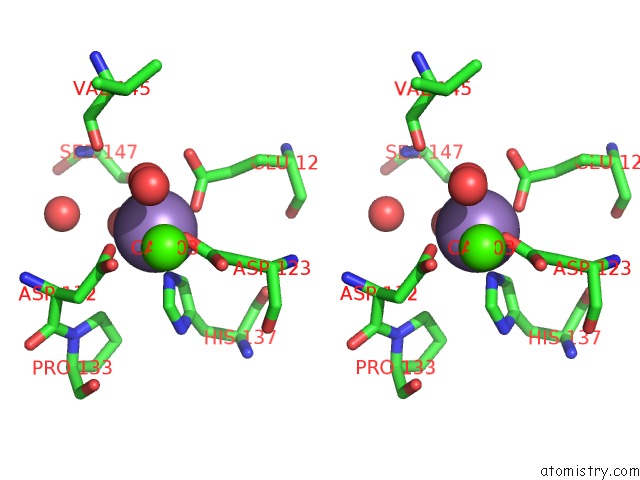
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Peanut Lectin Complexed with N-Beta-D-Galactopyranosyl-L-Succinamoyl Derivative (Ngs) within 5.0Å range:
|
Manganese binding site 3 out of 4 in 6vaw
Go back to
Manganese binding site 3 out
of 4 in the Peanut Lectin Complexed with N-Beta-D-Galactopyranosyl-L-Succinamoyl Derivative (Ngs)
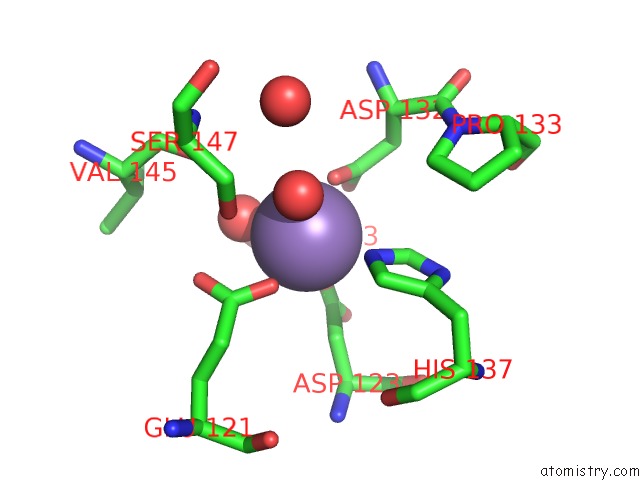
Mono view
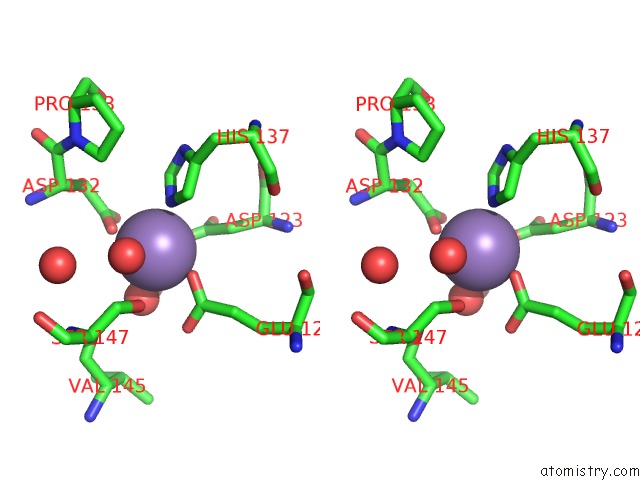
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Peanut Lectin Complexed with N-Beta-D-Galactopyranosyl-L-Succinamoyl Derivative (Ngs) within 5.0Å range:
|
Manganese binding site 4 out of 4 in 6vaw
Go back to
Manganese binding site 4 out
of 4 in the Peanut Lectin Complexed with N-Beta-D-Galactopyranosyl-L-Succinamoyl Derivative (Ngs)
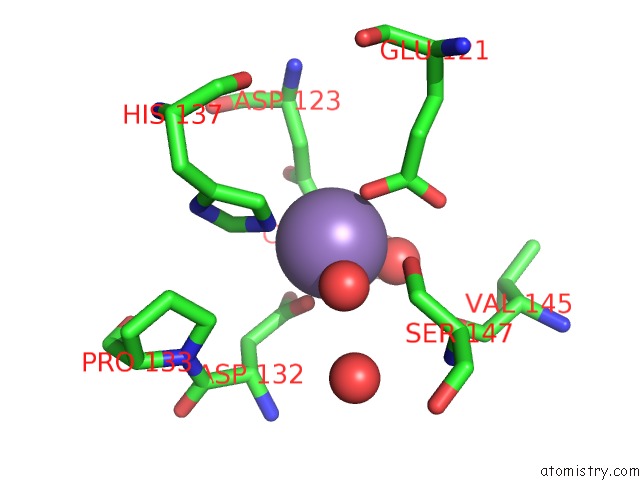
Mono view
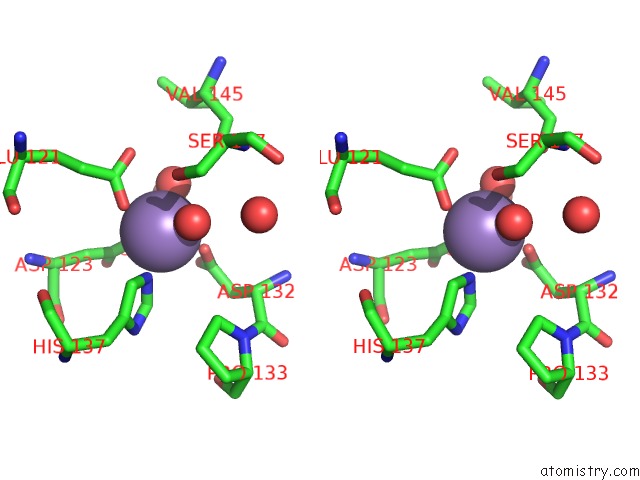
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of Peanut Lectin Complexed with N-Beta-D-Galactopyranosyl-L-Succinamoyl Derivative (Ngs) within 5.0Å range:
|
Reference:
A.J.Cagnoni,
E.D.Primo,
S.Klinke,
M.E.Cano,
W.Giordano,
K.V.Marino,
J.Kovensky,
F.A.Goldbaum,
M.L.Uhrig,
L.H.Otero.
Crystal Structures of Peanut Lectin in the Presence of Synthetic Beta-N- and Beta-S-Galactosides Disclose Evidence For the Recognition of Different Glycomimetic Ligands. Acta Crystallogr D Struct V. 76 1080 2020BIOL.
ISSN: ISSN 2059-7983
PubMed: 33135679
DOI: 10.1107/S2059798320012371
Page generated: Sun Oct 6 07:25:38 2024
ISSN: ISSN 2059-7983
PubMed: 33135679
DOI: 10.1107/S2059798320012371
Last articles
I in 5MWKI in 5MOZ
I in 5MSQ
I in 5M1Y
I in 5MPH
I in 5MJO
I in 5MHP
I in 5MA2
I in 5MHG
I in 5MHE