Manganese »
PDB 6rwz-6txf »
6sa1 »
Manganese in PDB 6sa1: Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp
Protein crystallography data
The structure of Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp, PDB code: 6sa1
was solved by
N.C.Brissett,
A.J.Doherty,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 88.66 / 2.01 |
| Space group | P 4 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 198.250, 198.250, 85.300, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.5 / 20.4 |
Other elements in 6sa1:
The structure of Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Manganese Binding Sites:
The binding sites of Manganese atom in the Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp
(pdb code 6sa1). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 4 binding sites of Manganese where determined in the Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp, PDB code: 6sa1:
Jump to Manganese binding site number: 1; 2; 3; 4;
In total 4 binding sites of Manganese where determined in the Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp, PDB code: 6sa1:
Jump to Manganese binding site number: 1; 2; 3; 4;
Manganese binding site 1 out of 4 in 6sa1
Go back to
Manganese binding site 1 out
of 4 in the Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp
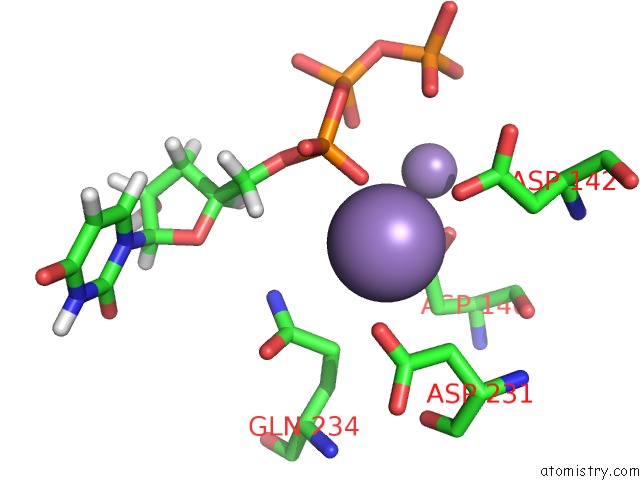
Mono view
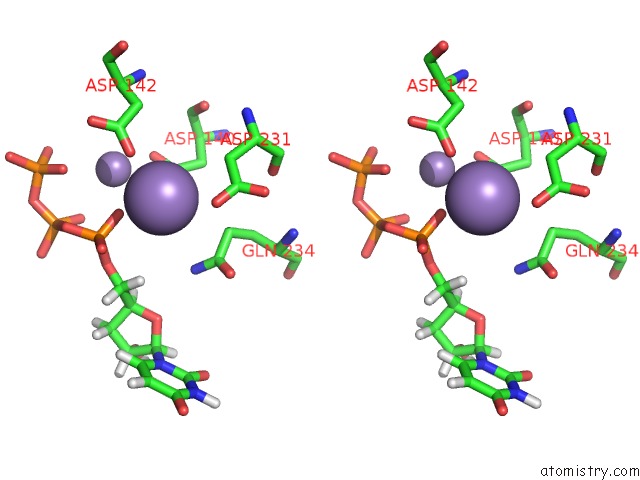
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp within 5.0Å range:
|
Manganese binding site 2 out of 4 in 6sa1
Go back to
Manganese binding site 2 out
of 4 in the Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp
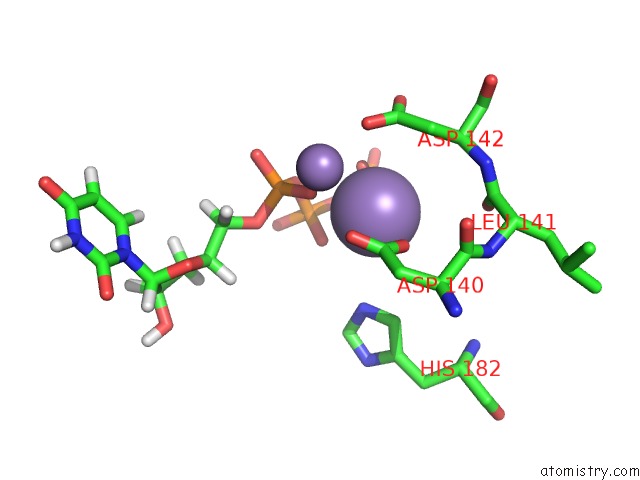
Mono view
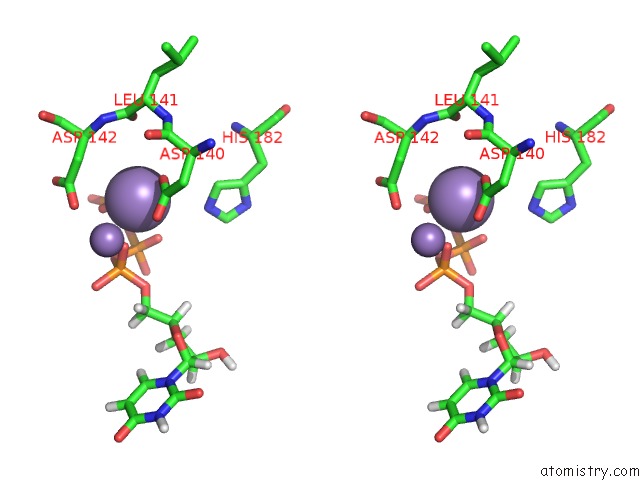
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp within 5.0Å range:
|
Manganese binding site 3 out of 4 in 6sa1
Go back to
Manganese binding site 3 out
of 4 in the Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp
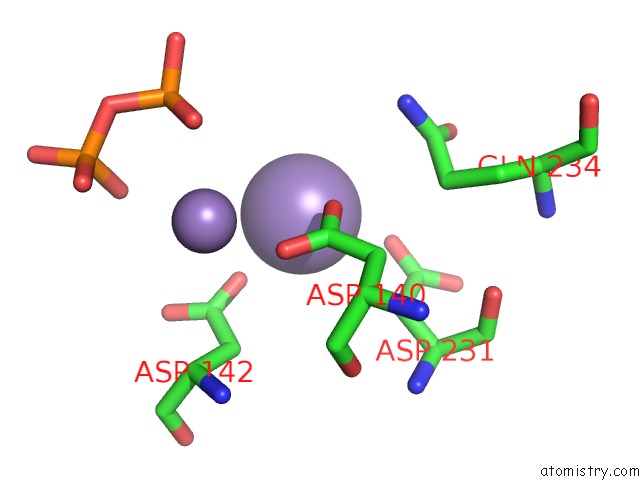
Mono view
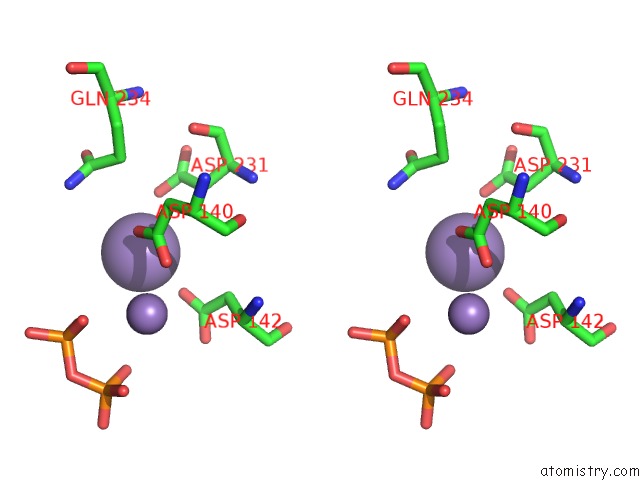
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp within 5.0Å range:
|
Manganese binding site 4 out of 4 in 6sa1
Go back to
Manganese binding site 4 out
of 4 in the Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp
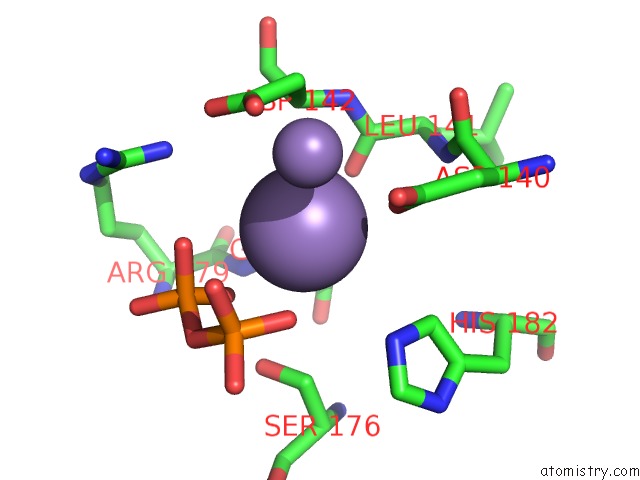
Mono view
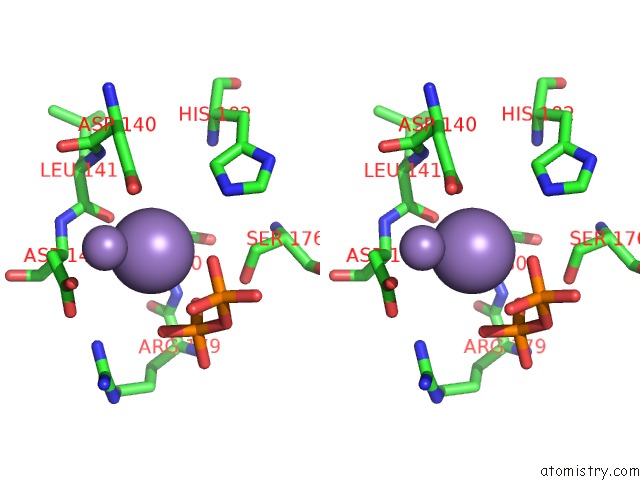
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp within 5.0Å range:
|
Reference:
N.C.Brissett,
K.Zabrady,
P.Plocinski,
J.Bianchi,
M.Korycka-Machala,
A.Brzostek,
J.Dziadek,
A.J.Doherty.
Molecular Basis For Dna Repair Synthesis on Short Gaps By Mycobacterial Primase-Polymerase C. Nat Commun V. 11 4196 2020.
ISSN: ESSN 2041-1723
PubMed: 32826907
DOI: 10.1038/S41467-020-18012-8
Page generated: Sun Oct 6 07:06:37 2024
ISSN: ESSN 2041-1723
PubMed: 32826907
DOI: 10.1038/S41467-020-18012-8
Last articles
I in 8H3ZI in 8H5L
I in 8II3
I in 8II1
I in 8IG1
I in 8GYR
I in 8GVV
I in 8GLI
I in 8GLH
I in 8EM5