Manganese »
PDB 6m30-6obu »
6nvo »
Manganese in PDB 6nvo: Crystal Structure of Pseudomonas Putida Nuclease Mpe
Protein crystallography data
The structure of Crystal Structure of Pseudomonas Putida Nuclease Mpe, PDB code: 6nvo
was solved by
Y.Goldgur,
S.Shuman,
A.Ejaz,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 36.50 / 2.20 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 58.224, 72.995, 41.748, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.2 / 26.3 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Crystal Structure of Pseudomonas Putida Nuclease Mpe
(pdb code 6nvo). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 2 binding sites of Manganese where determined in the Crystal Structure of Pseudomonas Putida Nuclease Mpe, PDB code: 6nvo:
Jump to Manganese binding site number: 1; 2;
In total 2 binding sites of Manganese where determined in the Crystal Structure of Pseudomonas Putida Nuclease Mpe, PDB code: 6nvo:
Jump to Manganese binding site number: 1; 2;
Manganese binding site 1 out of 2 in 6nvo
Go back to
Manganese binding site 1 out
of 2 in the Crystal Structure of Pseudomonas Putida Nuclease Mpe
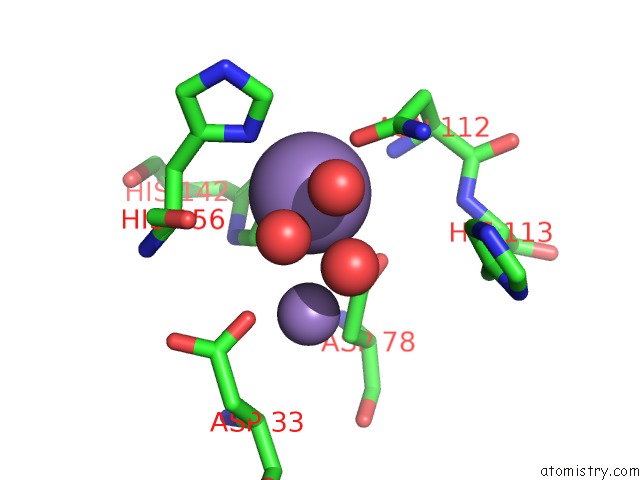
Mono view
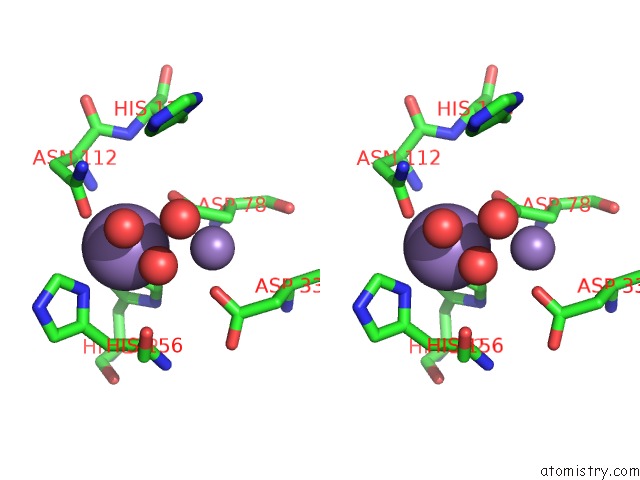
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Crystal Structure of Pseudomonas Putida Nuclease Mpe within 5.0Å range:
|
Manganese binding site 2 out of 2 in 6nvo
Go back to
Manganese binding site 2 out
of 2 in the Crystal Structure of Pseudomonas Putida Nuclease Mpe
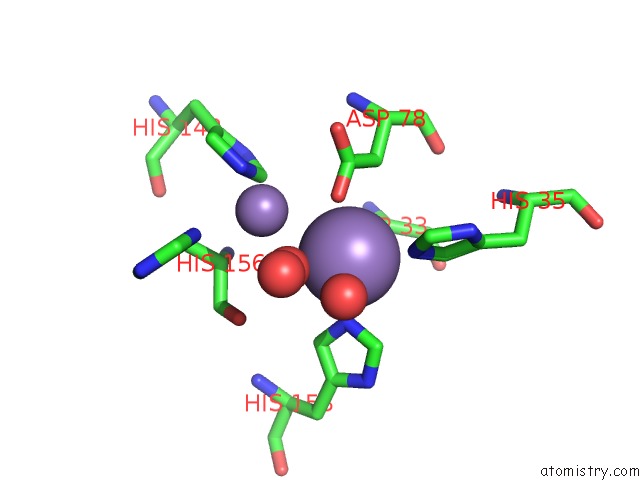
Mono view
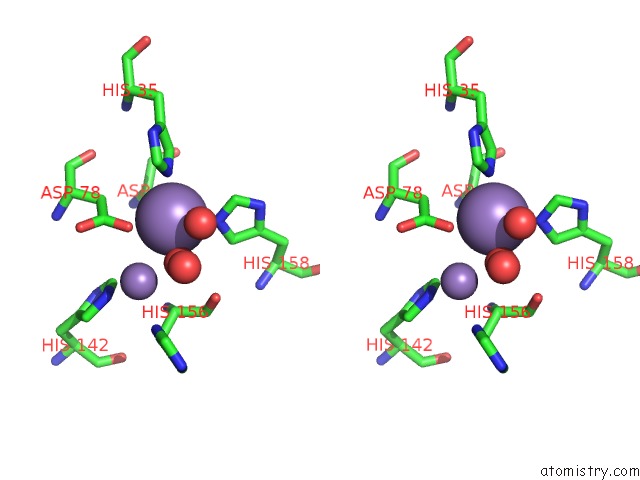
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Crystal Structure of Pseudomonas Putida Nuclease Mpe within 5.0Å range:
|
Reference:
A.Ejaz,
Y.Goldgur,
S.Shuman.
Activity and Structure Ofpseudomonas Putidampe, A Manganese-Dependent Single-Strand Dna Endonuclease Encoded in A Nucleic Acid Repair Gene Cluster. J.Biol.Chem. V. 294 7931 2019.
ISSN: ESSN 1083-351X
PubMed: 30894417
DOI: 10.1074/JBC.RA119.008049
Page generated: Sun Oct 6 05:38:05 2024
ISSN: ESSN 1083-351X
PubMed: 30894417
DOI: 10.1074/JBC.RA119.008049
Last articles
Mg in 6YRCMg in 6YRJ
Mg in 6YRD
Mg in 6YR8
Mg in 6YQ4
Mg in 6YR4
Mg in 6YPN
Mg in 6YOM
Mg in 6YL5
Mg in 6YP4