Manganese »
PDB 5z2k-6a9u »
5zee »
Manganese in PDB 5zee: Crystal Structure of Entamoeba Histolytica Arginase in Complex with N(Omega)-Hydroxy-L-Arginine (Noha) at 1.74 A
Enzymatic activity of Crystal Structure of Entamoeba Histolytica Arginase in Complex with N(Omega)-Hydroxy-L-Arginine (Noha) at 1.74 A
All present enzymatic activity of Crystal Structure of Entamoeba Histolytica Arginase in Complex with N(Omega)-Hydroxy-L-Arginine (Noha) at 1.74 A:
3.5.3.1;
3.5.3.1;
Protein crystallography data
The structure of Crystal Structure of Entamoeba Histolytica Arginase in Complex with N(Omega)-Hydroxy-L-Arginine (Noha) at 1.74 A, PDB code: 5zee
was solved by
A.Malik,
V.Dalal,
S.Ankri,
S.Tomar,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.14 / 1.74 |
| Space group | I 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 87.722, 97.538, 125.067, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.6 / 19.3 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Crystal Structure of Entamoeba Histolytica Arginase in Complex with N(Omega)-Hydroxy-L-Arginine (Noha) at 1.74 A
(pdb code 5zee). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 4 binding sites of Manganese where determined in the Crystal Structure of Entamoeba Histolytica Arginase in Complex with N(Omega)-Hydroxy-L-Arginine (Noha) at 1.74 A, PDB code: 5zee:
Jump to Manganese binding site number: 1; 2; 3; 4;
In total 4 binding sites of Manganese where determined in the Crystal Structure of Entamoeba Histolytica Arginase in Complex with N(Omega)-Hydroxy-L-Arginine (Noha) at 1.74 A, PDB code: 5zee:
Jump to Manganese binding site number: 1; 2; 3; 4;
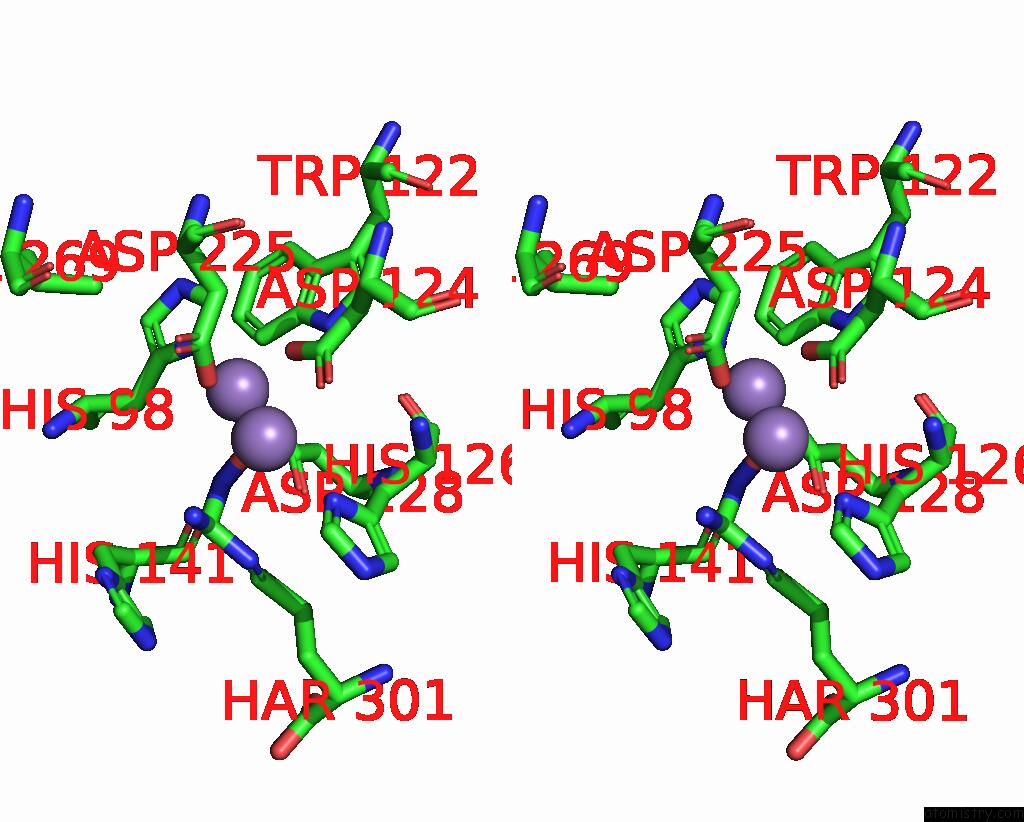
Manganese binding site 1 out of 4 in 5zee
Go back to
Manganese binding site 1 out
of 4 in the Crystal Structure of Entamoeba Histolytica Arginase in Complex with N(Omega)-Hydroxy-L-Arginine (Noha) at 1.74 A

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Crystal Structure of Entamoeba Histolytica Arginase in Complex with N(Omega)-Hydroxy-L-Arginine (Noha) at 1.74 A within 5.0Å range:
|
Manganese binding site 2 out of 4 in 5zee
Go back to
Manganese binding site 2 out
of 4 in the Crystal Structure of Entamoeba Histolytica Arginase in Complex with N(Omega)-Hydroxy-L-Arginine (Noha) at 1.74 A

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Crystal Structure of Entamoeba Histolytica Arginase in Complex with N(Omega)-Hydroxy-L-Arginine (Noha) at 1.74 A within 5.0Å range:
|
Manganese binding site 3 out of 4 in 5zee
Go back to
Manganese binding site 3 out
of 4 in the Crystal Structure of Entamoeba Histolytica Arginase in Complex with N(Omega)-Hydroxy-L-Arginine (Noha) at 1.74 A

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Crystal Structure of Entamoeba Histolytica Arginase in Complex with N(Omega)-Hydroxy-L-Arginine (Noha) at 1.74 A within 5.0Å range:
|
Manganese binding site 4 out of 4 in 5zee
Go back to
Manganese binding site 4 out
of 4 in the Crystal Structure of Entamoeba Histolytica Arginase in Complex with N(Omega)-Hydroxy-L-Arginine (Noha) at 1.74 A

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of Crystal Structure of Entamoeba Histolytica Arginase in Complex with N(Omega)-Hydroxy-L-Arginine (Noha) at 1.74 A within 5.0Å range:
|
Reference:
A.Malik,
V.Dalal,
S.Ankri,
S.Tomar.
Structural Insights Into Entamoeba Histolytica Arginase and Structure-Based Identification of Novel Non-Amino Acid Based Inhibitors As Potential Antiamoebic Molecules. Febs J. V. 286 4135 2019.
ISSN: ISSN 1742-464X
PubMed: 31199070
DOI: 10.1111/FEBS.14960
Page generated: Sun Oct 6 03:40:01 2024
ISSN: ISSN 1742-464X
PubMed: 31199070
DOI: 10.1111/FEBS.14960
Last articles
K in 8K1EK in 8JZG
K in 8K0U
K in 8K0T
K in 8JGW
K in 8JZA
K in 8JC7
K in 8JGR
K in 8IYM
K in 8JAH