Manganese »
PDB 4wl8-4xwt »
4xsl »
Manganese in PDB 4xsl: Crystal Strcutre of D-Tagatose 3-Epimerase C66S From Pseudomonas Cichorii in Complex with Glycerol
Protein crystallography data
The structure of Crystal Strcutre of D-Tagatose 3-Epimerase C66S From Pseudomonas Cichorii in Complex with Glycerol, PDB code: 4xsl
was solved by
H.Yoshida,
A.Yoshihara,
T.Ishii,
K.Izumori,
S.Kamitori,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 32.07 / 1.60 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 52.271, 124.552, 93.319, 90.00, 98.74, 90.00 |
| R / Rfree (%) | 17.8 / 20.5 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Crystal Strcutre of D-Tagatose 3-Epimerase C66S From Pseudomonas Cichorii in Complex with Glycerol
(pdb code 4xsl). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 4 binding sites of Manganese where determined in the Crystal Strcutre of D-Tagatose 3-Epimerase C66S From Pseudomonas Cichorii in Complex with Glycerol, PDB code: 4xsl:
Jump to Manganese binding site number: 1; 2; 3; 4;
In total 4 binding sites of Manganese where determined in the Crystal Strcutre of D-Tagatose 3-Epimerase C66S From Pseudomonas Cichorii in Complex with Glycerol, PDB code: 4xsl:
Jump to Manganese binding site number: 1; 2; 3; 4;
Manganese binding site 1 out of 4 in 4xsl
Go back to
Manganese binding site 1 out
of 4 in the Crystal Strcutre of D-Tagatose 3-Epimerase C66S From Pseudomonas Cichorii in Complex with Glycerol
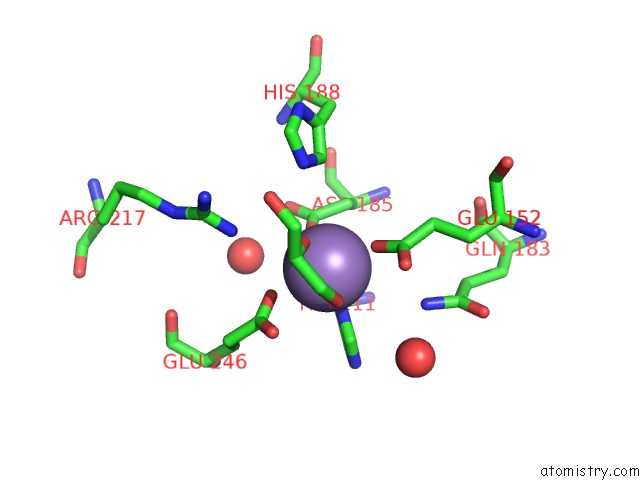
Mono view
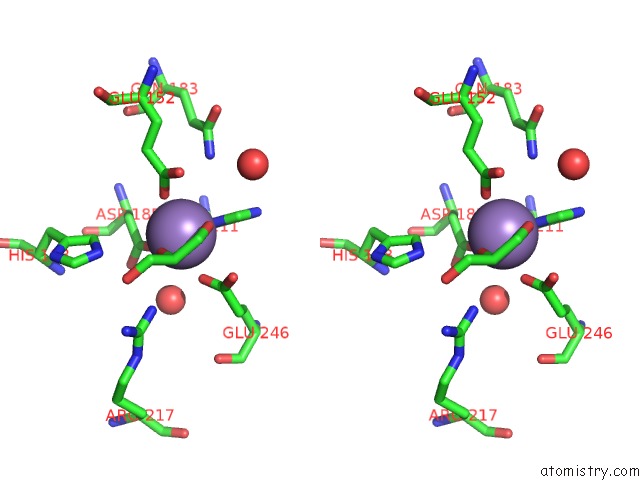
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Crystal Strcutre of D-Tagatose 3-Epimerase C66S From Pseudomonas Cichorii in Complex with Glycerol within 5.0Å range:
|
Manganese binding site 2 out of 4 in 4xsl
Go back to
Manganese binding site 2 out
of 4 in the Crystal Strcutre of D-Tagatose 3-Epimerase C66S From Pseudomonas Cichorii in Complex with Glycerol
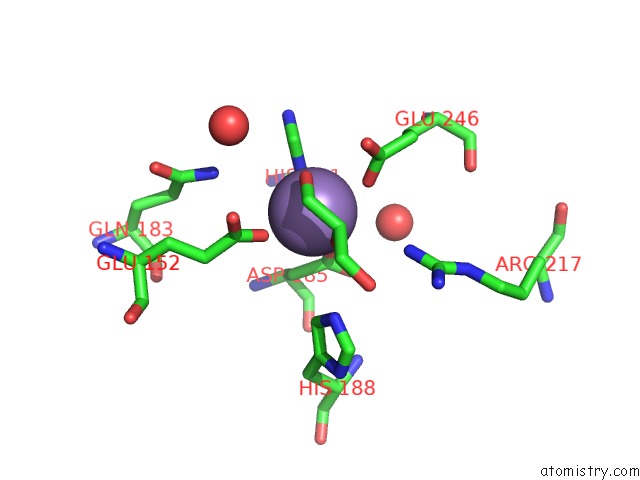
Mono view
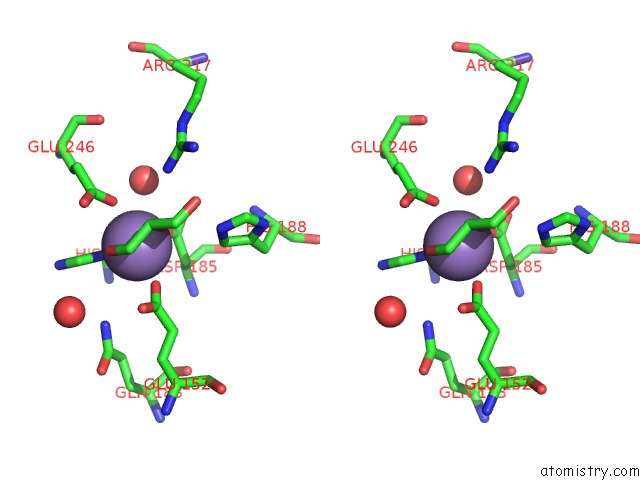
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Crystal Strcutre of D-Tagatose 3-Epimerase C66S From Pseudomonas Cichorii in Complex with Glycerol within 5.0Å range:
|
Manganese binding site 3 out of 4 in 4xsl
Go back to
Manganese binding site 3 out
of 4 in the Crystal Strcutre of D-Tagatose 3-Epimerase C66S From Pseudomonas Cichorii in Complex with Glycerol
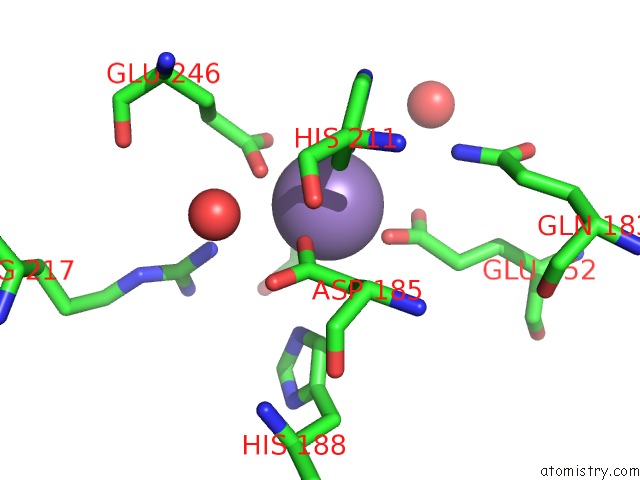
Mono view
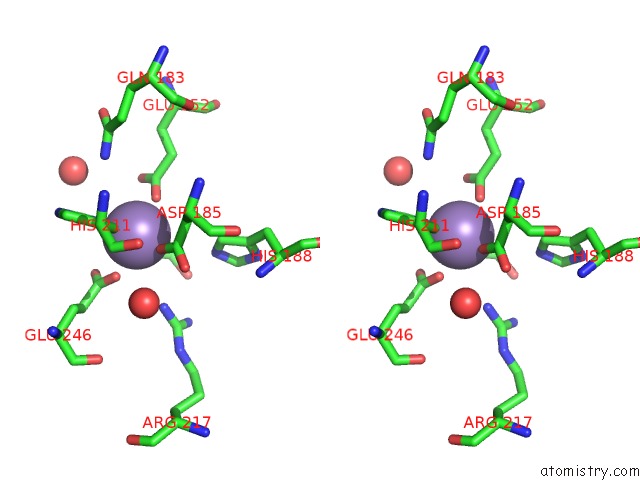
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Crystal Strcutre of D-Tagatose 3-Epimerase C66S From Pseudomonas Cichorii in Complex with Glycerol within 5.0Å range:
|
Manganese binding site 4 out of 4 in 4xsl
Go back to
Manganese binding site 4 out
of 4 in the Crystal Strcutre of D-Tagatose 3-Epimerase C66S From Pseudomonas Cichorii in Complex with Glycerol
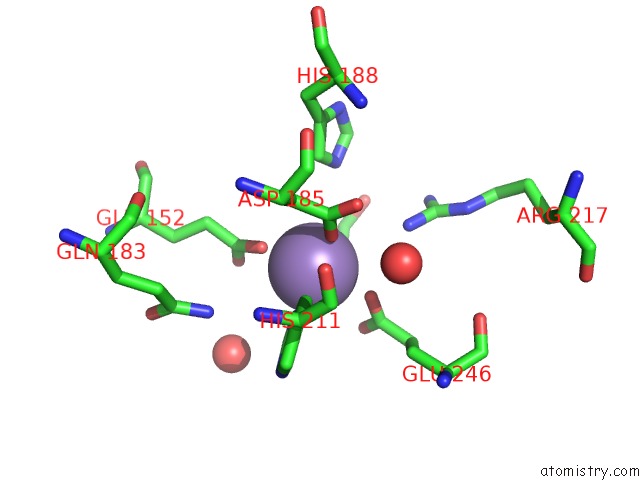
Mono view
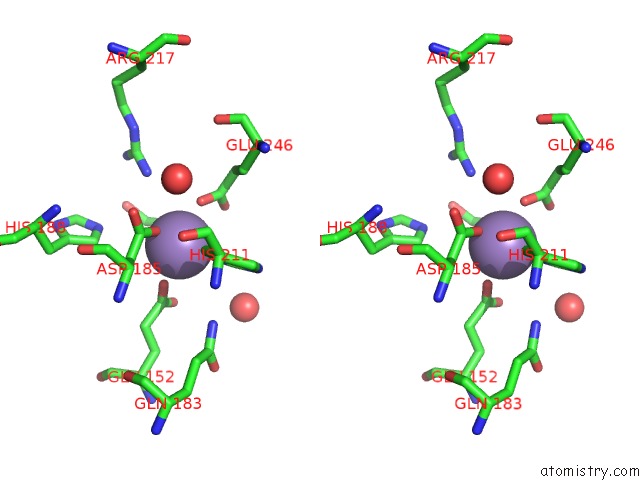
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of Crystal Strcutre of D-Tagatose 3-Epimerase C66S From Pseudomonas Cichorii in Complex with Glycerol within 5.0Å range:
|
Reference:
H.Yoshida,
A.Yoshihara,
T.Ishii,
K.Izumori,
S.Kamitori.
X-Ray Structures of the Pseudomonas Cichorii D-Tagatose 3-Epimerase Mutant Form C66S Recognizing Deoxy Sugars As Substrates Appl. Microbiol. Biotechnol. V. 100 10403 2016.
ISSN: ESSN 1432-0614
PubMed: 27368739
DOI: 10.1007/S00253-016-7673-7
Page generated: Sat Oct 5 22:56:10 2024
ISSN: ESSN 1432-0614
PubMed: 27368739
DOI: 10.1007/S00253-016-7673-7
Last articles
K in 2ZD9K in 3A3U
K in 2ZWU
K in 2YIA
K in 2ZWT
K in 2ZUJ
K in 2ZUI
K in 2ZUH
K in 2ZAX
K in 2ZAW