Manganese »
PDB 4b5i-4dec »
4cug »
Manganese in PDB 4cug: Rhodothermus Marinus Ycfd-Like Ribosomal Protein L16 Arginyl Hydroxylase in Complex Substrate Fragment
Protein crystallography data
The structure of Rhodothermus Marinus Ycfd-Like Ribosomal Protein L16 Arginyl Hydroxylase in Complex Substrate Fragment, PDB code: 4cug
was solved by
M.A.Mcdonough,
R.Sekirnik,
C.J.Schofield,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 58.74 / 2.96 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 98.220, 117.470, 120.280, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.01 / 22.7 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Rhodothermus Marinus Ycfd-Like Ribosomal Protein L16 Arginyl Hydroxylase in Complex Substrate Fragment
(pdb code 4cug). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 2 binding sites of Manganese where determined in the Rhodothermus Marinus Ycfd-Like Ribosomal Protein L16 Arginyl Hydroxylase in Complex Substrate Fragment, PDB code: 4cug:
Jump to Manganese binding site number: 1; 2;
In total 2 binding sites of Manganese where determined in the Rhodothermus Marinus Ycfd-Like Ribosomal Protein L16 Arginyl Hydroxylase in Complex Substrate Fragment, PDB code: 4cug:
Jump to Manganese binding site number: 1; 2;
Manganese binding site 1 out of 2 in 4cug
Go back to
Manganese binding site 1 out
of 2 in the Rhodothermus Marinus Ycfd-Like Ribosomal Protein L16 Arginyl Hydroxylase in Complex Substrate Fragment
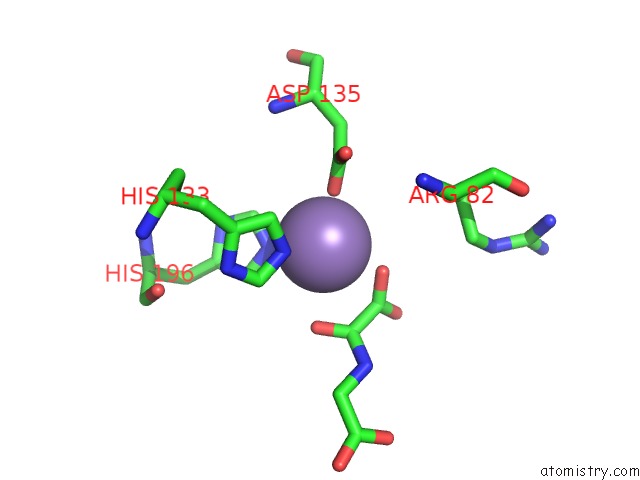
Mono view
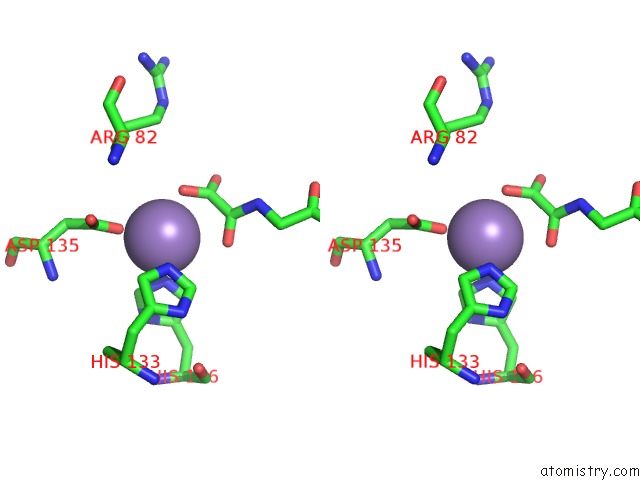
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Rhodothermus Marinus Ycfd-Like Ribosomal Protein L16 Arginyl Hydroxylase in Complex Substrate Fragment within 5.0Å range:
|
Manganese binding site 2 out of 2 in 4cug
Go back to
Manganese binding site 2 out
of 2 in the Rhodothermus Marinus Ycfd-Like Ribosomal Protein L16 Arginyl Hydroxylase in Complex Substrate Fragment
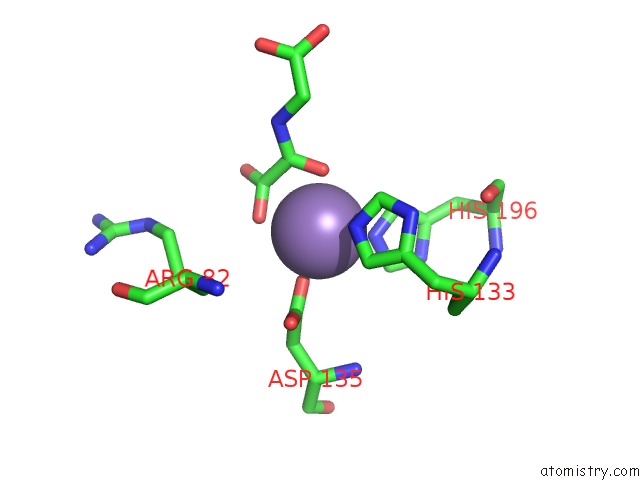
Mono view
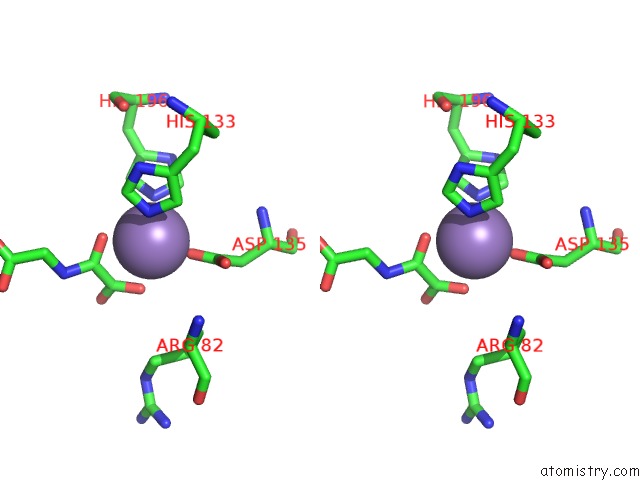
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Rhodothermus Marinus Ycfd-Like Ribosomal Protein L16 Arginyl Hydroxylase in Complex Substrate Fragment within 5.0Å range:
|
Reference:
R.Chowdhury,
R.Sekirnik,
N.C.Brissett,
T.Krojer,
C.-H.Ho,
S.S.Ng,
I.J.Clifton,
W.Ge,
N.J.Kershaw,
G.C.Fox,
J.R.C.Muniz,
M.Vollmar,
C.Phillips,
E.S.Pilka,
K.L.Kavanagh,
F.Von Deflt,
U.Oppermann,
M.A.Mcdonough,
A.J.Doherty,
C.J.Schofield.
Ribosomal Oxygenases Are Structurally Conserved From Prokaryotes to Humans. Nature V. 510 422 2014.
ISSN: ISSN 0028-0836
PubMed: 24814345
DOI: 10.1038/NATURE13263
Page generated: Sat Oct 5 18:55:58 2024
ISSN: ISSN 0028-0836
PubMed: 24814345
DOI: 10.1038/NATURE13263
Last articles
K in 6CF1K in 6CI0
K in 6C9X
K in 6C65
K in 6C64
K in 6C9U
K in 6C63
K in 6C0Y
K in 6C3O
K in 6C3Z