Manganese »
PDB 3uag-3vnm »
3vnm »
Manganese in PDB 3vnm: Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10
Protein crystallography data
The structure of Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10, PDB code: 3vnm
was solved by
H.C.Chan,
Y.Zhu,
Y.Hu,
T.P.Ko,
C.H.Huang,
F.Ren,
C.C.Chen,
R.T.Guo,
Y.Sun,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.00 / 2.12 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 79.924, 115.225, 91.614, 90.00, 105.73, 90.00 |
| R / Rfree (%) | 19 / 23.2 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10
(pdb code 3vnm). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 8 binding sites of Manganese where determined in the Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10, PDB code: 3vnm:
Jump to Manganese binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Manganese where determined in the Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10, PDB code: 3vnm:
Jump to Manganese binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
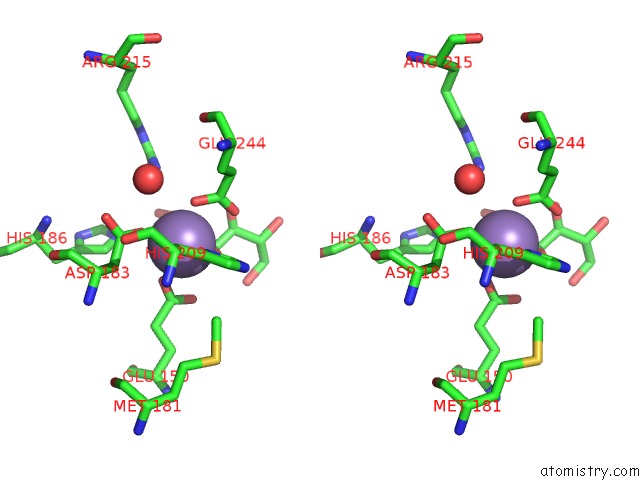
Manganese binding site 1 out of 8 in 3vnm
Go back to
Manganese binding site 1 out
of 8 in the Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10 within 5.0Å range:
|
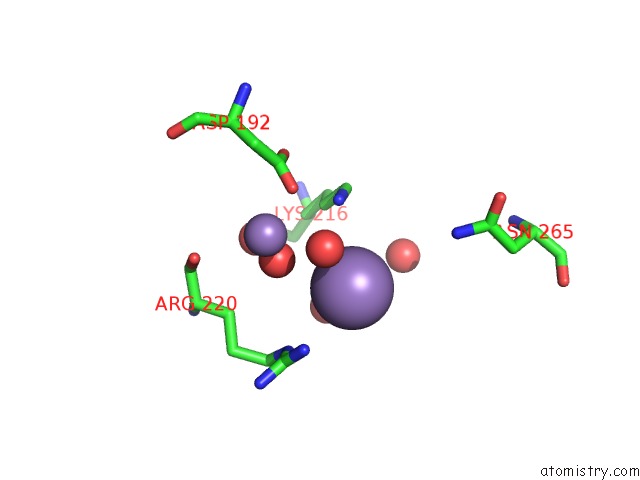
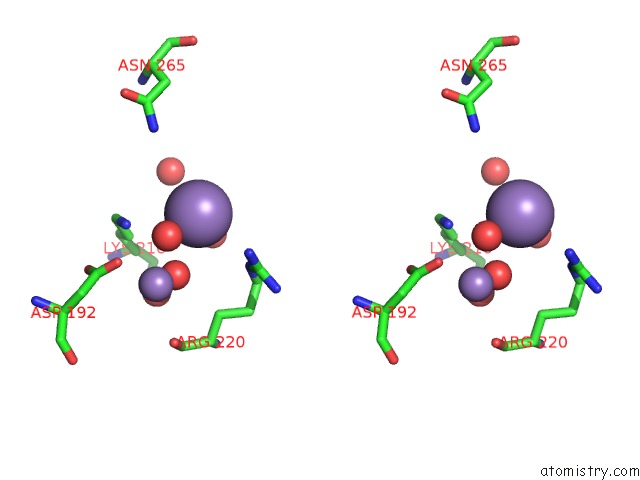
Manganese binding site 2 out of 8 in 3vnm
Go back to
Manganese binding site 2 out
of 8 in the Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10 within 5.0Å range:
|
Manganese binding site 3 out of 8 in 3vnm
Go back to
Manganese binding site 3 out
of 8 in the Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10 within 5.0Å range:
|
Manganese binding site 4 out of 8 in 3vnm
Go back to
Manganese binding site 4 out
of 8 in the Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10 within 5.0Å range:
|
Manganese binding site 5 out of 8 in 3vnm
Go back to
Manganese binding site 5 out
of 8 in the Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 5 of Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10 within 5.0Å range:
|
Manganese binding site 6 out of 8 in 3vnm
Go back to
Manganese binding site 6 out
of 8 in the Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 6 of Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10 within 5.0Å range:
|
Manganese binding site 7 out of 8 in 3vnm
Go back to
Manganese binding site 7 out
of 8 in the Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 7 of Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10 within 5.0Å range:
|
Manganese binding site 8 out of 8 in 3vnm
Go back to
Manganese binding site 8 out
of 8 in the Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 8 of Crystal Structures of D-Psicose 3-Epimerase with D-Sorbose From Clostridium Cellulolyticum H10 within 5.0Å range:
|
Reference:
H.C.Chan,
Y.Zhu,
Y.Hu,
T.P.Ko,
C.H.Huang,
F.Ren,
C.C.Chen,
Y.Ma,
R.T.Guo,
Y.Sun.
Crystal Structures of D-Psicose 3-Epimerase From Clostridium Cellulolyticum H10 and Its Complex with Ketohexose Sugars. Protein Cell V. 3 123 2012.
ISSN: ISSN 1674-800X
PubMed: 22426981
DOI: 10.1007/S13238-012-2026-5
Page generated: Sat Oct 5 18:21:40 2024
ISSN: ISSN 1674-800X
PubMed: 22426981
DOI: 10.1007/S13238-012-2026-5
Last articles
K in 3BWPK in 3BOL
K in 3BOF
K in 3BWM
K in 3BEH
K in 3BNS
K in 3BO3
K in 3BJF
K in 3BNR
K in 3BNQ