Manganese »
PDB 3q7c-3rl4 »
3ras »
Manganese in PDB 3ras: Crystal Structure of 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase (Dxr) Complexed with A Lipophilic Phosphonate Inhibitor
Enzymatic activity of Crystal Structure of 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase (Dxr) Complexed with A Lipophilic Phosphonate Inhibitor
All present enzymatic activity of Crystal Structure of 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase (Dxr) Complexed with A Lipophilic Phosphonate Inhibitor:
1.1.1.267;
1.1.1.267;
Protein crystallography data
The structure of Crystal Structure of 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase (Dxr) Complexed with A Lipophilic Phosphonate Inhibitor, PDB code: 3ras
was solved by
J.Diao,
L.Deng,
B.V.V.Prasad,
Y.Song,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 33.45 / 2.55 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 67.250, 65.179, 85.408, 90.00, 101.28, 90.00 |
| R / Rfree (%) | 20.8 / 27.8 |
Other elements in 3ras:
The structure of Crystal Structure of 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase (Dxr) Complexed with A Lipophilic Phosphonate Inhibitor also contains other interesting chemical elements:
| Chlorine | (Cl) | 8 atoms |
Manganese Binding Sites:
The binding sites of Manganese atom in the Crystal Structure of 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase (Dxr) Complexed with A Lipophilic Phosphonate Inhibitor
(pdb code 3ras). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 2 binding sites of Manganese where determined in the Crystal Structure of 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase (Dxr) Complexed with A Lipophilic Phosphonate Inhibitor, PDB code: 3ras:
Jump to Manganese binding site number: 1; 2;
In total 2 binding sites of Manganese where determined in the Crystal Structure of 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase (Dxr) Complexed with A Lipophilic Phosphonate Inhibitor, PDB code: 3ras:
Jump to Manganese binding site number: 1; 2;
Manganese binding site 1 out of 2 in 3ras
Go back to
Manganese binding site 1 out
of 2 in the Crystal Structure of 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase (Dxr) Complexed with A Lipophilic Phosphonate Inhibitor

Mono view
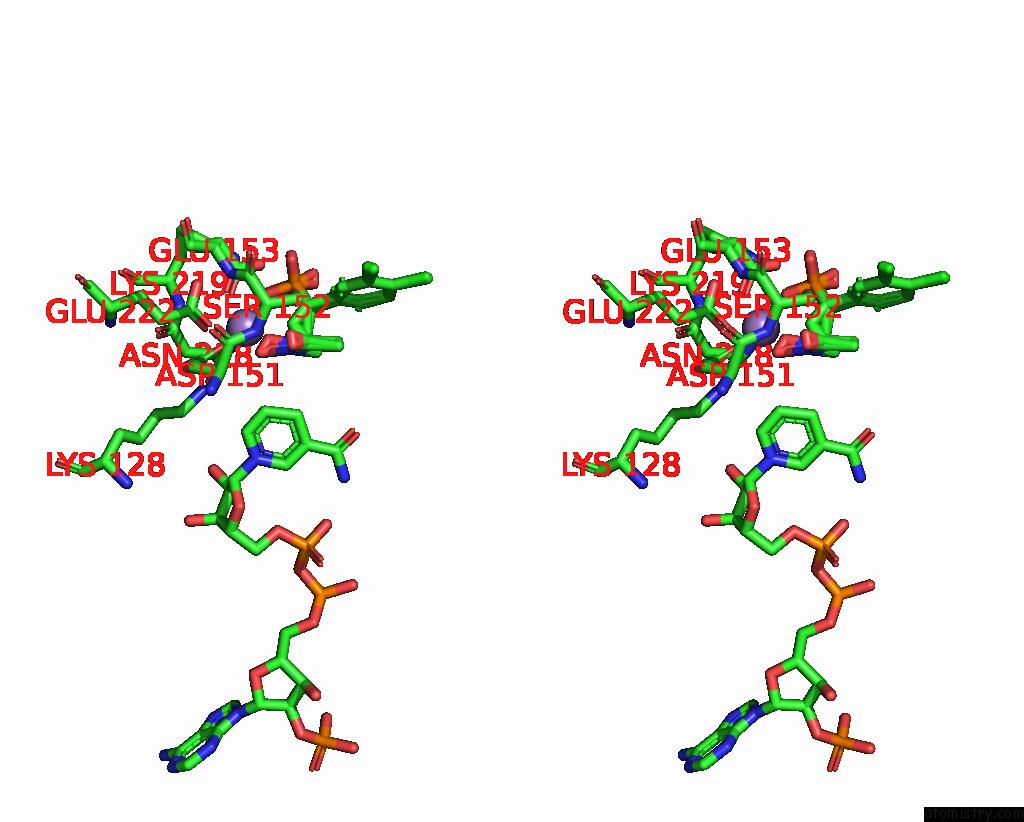
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Crystal Structure of 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase (Dxr) Complexed with A Lipophilic Phosphonate Inhibitor within 5.0Å range:
|
Manganese binding site 2 out of 2 in 3ras
Go back to
Manganese binding site 2 out
of 2 in the Crystal Structure of 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase (Dxr) Complexed with A Lipophilic Phosphonate Inhibitor
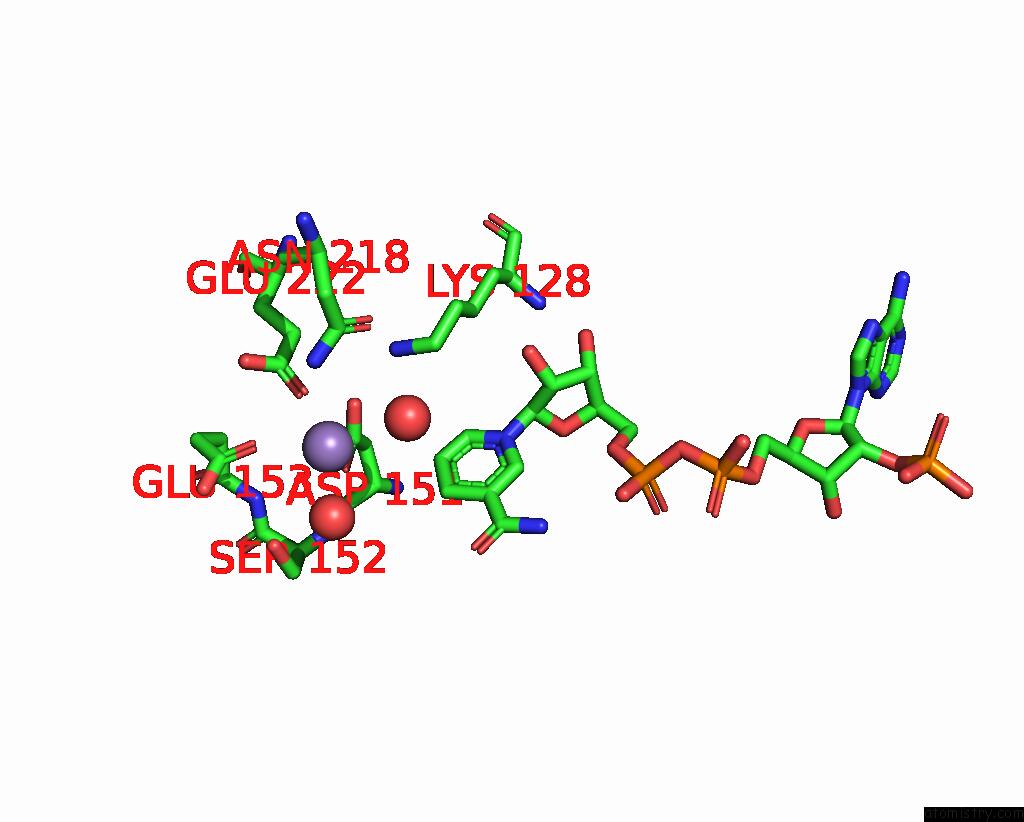
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Crystal Structure of 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase (Dxr) Complexed with A Lipophilic Phosphonate Inhibitor within 5.0Å range:
|
Reference:
L.Deng,
J.Diao,
P.Chen,
V.Pujari,
Y.Yao,
G.Cheng,
D.C.Crick,
B.V.Prasad,
Y.Song.
Inhibition of 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase By Lipophilic Phosphonates: Sar, Qsar, and Crystallographic Studies. J.Med.Chem. V. 54 4721 2011.
ISSN: ISSN 0022-2623
PubMed: 21561155
DOI: 10.1021/JM200363D
Page generated: Sat Oct 5 17:43:54 2024
ISSN: ISSN 0022-2623
PubMed: 21561155
DOI: 10.1021/JM200363D
Last articles
K in 4G38K in 4G0F
K in 4FXF
K in 4G1V
K in 4FXM
K in 4FB0
K in 4FXS
K in 4FO4
K in 4FAW
K in 4FWJ