Manganese »
PDB 1yw7-2a2t »
1zqm »
Manganese in PDB 1zqm: Dna Polymerase Beta (Pol B) (E.C.2.7.7.7) Complexed with Seven Base Pairs of Dna; Soaked in the Presence of MNCL2 (15 Millimolar)
Protein crystallography data
The structure of Dna Polymerase Beta (Pol B) (E.C.2.7.7.7) Complexed with Seven Base Pairs of Dna; Soaked in the Presence of MNCL2 (15 Millimolar), PDB code: 1zqm
was solved by
H.Pelletier,
M.R.Sawaya,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 3.20 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 178.881, 57.634, 48.080, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.9 / n/a |
Manganese Binding Sites:
The binding sites of Manganese atom in the Dna Polymerase Beta (Pol B) (E.C.2.7.7.7) Complexed with Seven Base Pairs of Dna; Soaked in the Presence of MNCL2 (15 Millimolar)
(pdb code 1zqm). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 3 binding sites of Manganese where determined in the Dna Polymerase Beta (Pol B) (E.C.2.7.7.7) Complexed with Seven Base Pairs of Dna; Soaked in the Presence of MNCL2 (15 Millimolar), PDB code: 1zqm:
Jump to Manganese binding site number: 1; 2; 3;
In total 3 binding sites of Manganese where determined in the Dna Polymerase Beta (Pol B) (E.C.2.7.7.7) Complexed with Seven Base Pairs of Dna; Soaked in the Presence of MNCL2 (15 Millimolar), PDB code: 1zqm:
Jump to Manganese binding site number: 1; 2; 3;
Manganese binding site 1 out of 3 in 1zqm
Go back to
Manganese binding site 1 out
of 3 in the Dna Polymerase Beta (Pol B) (E.C.2.7.7.7) Complexed with Seven Base Pairs of Dna; Soaked in the Presence of MNCL2 (15 Millimolar)
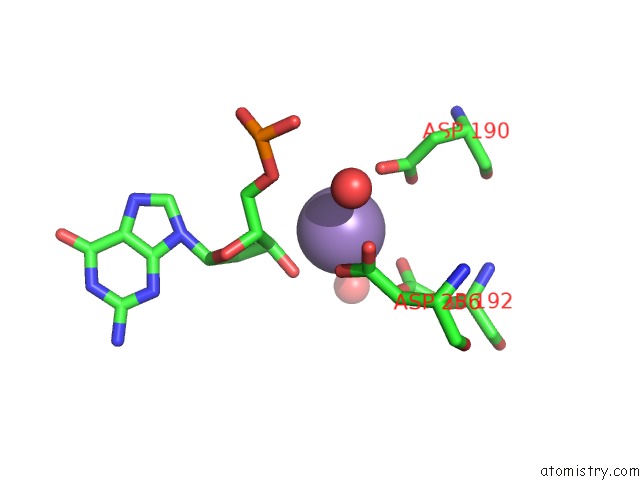
Mono view
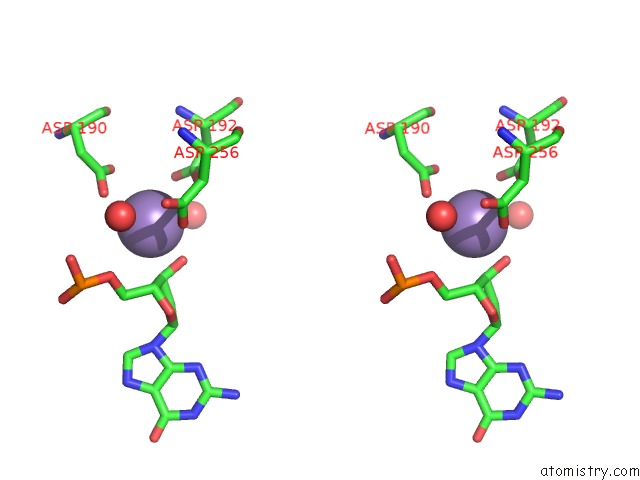
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Dna Polymerase Beta (Pol B) (E.C.2.7.7.7) Complexed with Seven Base Pairs of Dna; Soaked in the Presence of MNCL2 (15 Millimolar) within 5.0Å range:
|
Manganese binding site 2 out of 3 in 1zqm
Go back to
Manganese binding site 2 out
of 3 in the Dna Polymerase Beta (Pol B) (E.C.2.7.7.7) Complexed with Seven Base Pairs of Dna; Soaked in the Presence of MNCL2 (15 Millimolar)
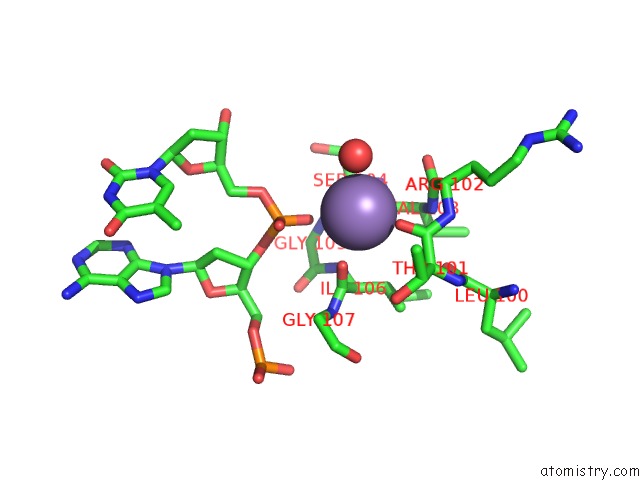
Mono view
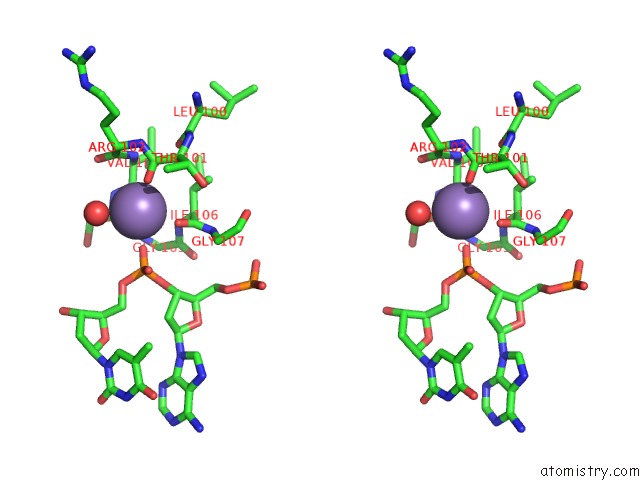
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Dna Polymerase Beta (Pol B) (E.C.2.7.7.7) Complexed with Seven Base Pairs of Dna; Soaked in the Presence of MNCL2 (15 Millimolar) within 5.0Å range:
|
Manganese binding site 3 out of 3 in 1zqm
Go back to
Manganese binding site 3 out
of 3 in the Dna Polymerase Beta (Pol B) (E.C.2.7.7.7) Complexed with Seven Base Pairs of Dna; Soaked in the Presence of MNCL2 (15 Millimolar)
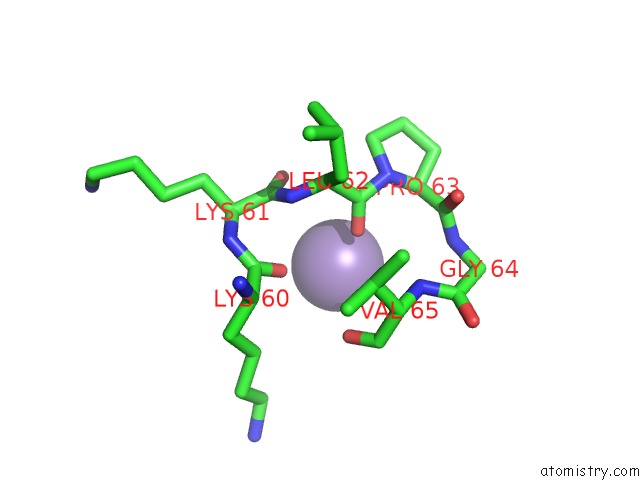
Mono view
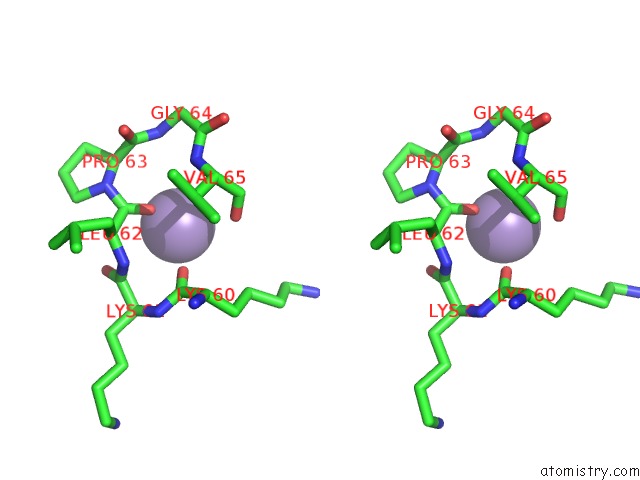
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Dna Polymerase Beta (Pol B) (E.C.2.7.7.7) Complexed with Seven Base Pairs of Dna; Soaked in the Presence of MNCL2 (15 Millimolar) within 5.0Å range:
|
Reference:
H.Pelletier,
M.R.Sawaya.
Characterization of the Metal Ion Binding Helix-Hairpin-Helix Motifs in Human Dna Polymerase Beta By X-Ray Structural Analysis. Biochemistry V. 35 12778 1996.
ISSN: ISSN 0006-2960
PubMed: 8841120
DOI: 10.1021/BI960790I
Page generated: Sat Oct 5 13:18:28 2024
ISSN: ISSN 0006-2960
PubMed: 8841120
DOI: 10.1021/BI960790I
Last articles
K in 4U5MK in 4U6C
K in 4U69
K in 4TWK
K in 4TS2
K in 4TOH
K in 4TOE
K in 4TOG
K in 4TOF
K in 4TOD