Manganese »
PDB 1o9i-1pj3 »
1p3d »
Manganese in PDB 1p3d: Crystal Structure of Udp-N-Acetylmuramic Acid:L-Alanine Ligase (Murc) in Complex with Uma and Anp.
Enzymatic activity of Crystal Structure of Udp-N-Acetylmuramic Acid:L-Alanine Ligase (Murc) in Complex with Uma and Anp.
All present enzymatic activity of Crystal Structure of Udp-N-Acetylmuramic Acid:L-Alanine Ligase (Murc) in Complex with Uma and Anp.:
6.3.2.8;
6.3.2.8;
Protein crystallography data
The structure of Crystal Structure of Udp-N-Acetylmuramic Acid:L-Alanine Ligase (Murc) in Complex with Uma and Anp., PDB code: 1p3d
was solved by
C.D.Mol,
A.Brooun,
D.R.Dougan,
M.T.Hilgers,
L.W.Tari,
R.A.Wijnands,
M.W.Knuth,
D.E.Mcree,
R.V.Swanson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.00 / 1.70 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 74.920, 87.328, 86.113, 90.00, 104.86, 90.00 |
| R / Rfree (%) | 16.8 / 19.4 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Crystal Structure of Udp-N-Acetylmuramic Acid:L-Alanine Ligase (Murc) in Complex with Uma and Anp.
(pdb code 1p3d). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 4 binding sites of Manganese where determined in the Crystal Structure of Udp-N-Acetylmuramic Acid:L-Alanine Ligase (Murc) in Complex with Uma and Anp., PDB code: 1p3d:
Jump to Manganese binding site number: 1; 2; 3; 4;
In total 4 binding sites of Manganese where determined in the Crystal Structure of Udp-N-Acetylmuramic Acid:L-Alanine Ligase (Murc) in Complex with Uma and Anp., PDB code: 1p3d:
Jump to Manganese binding site number: 1; 2; 3; 4;
Manganese binding site 1 out of 4 in 1p3d
Go back to
Manganese binding site 1 out
of 4 in the Crystal Structure of Udp-N-Acetylmuramic Acid:L-Alanine Ligase (Murc) in Complex with Uma and Anp.
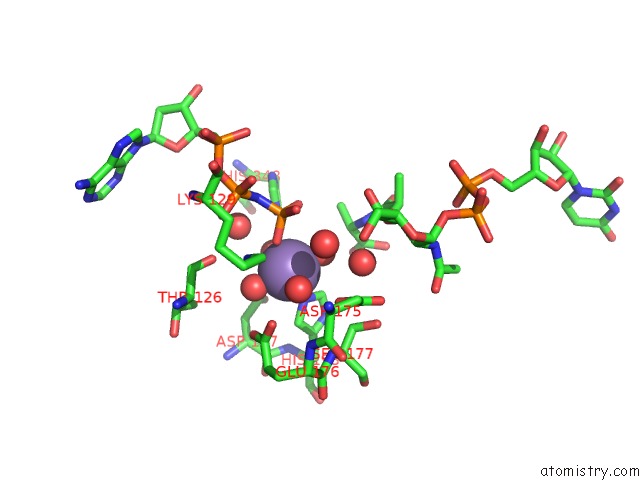
Mono view
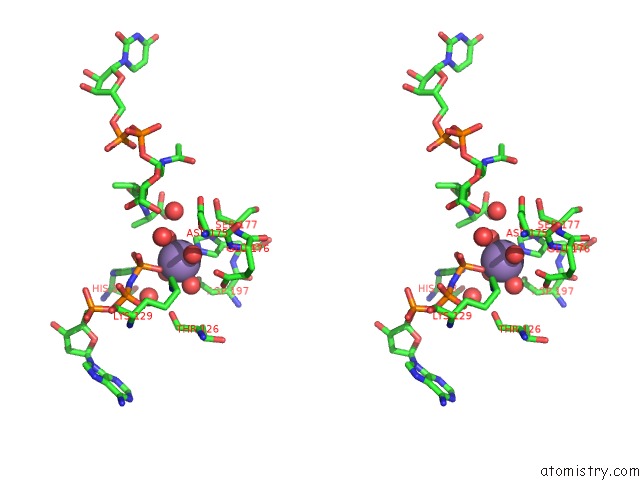
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Crystal Structure of Udp-N-Acetylmuramic Acid:L-Alanine Ligase (Murc) in Complex with Uma and Anp. within 5.0Å range:
|
Manganese binding site 2 out of 4 in 1p3d
Go back to
Manganese binding site 2 out
of 4 in the Crystal Structure of Udp-N-Acetylmuramic Acid:L-Alanine Ligase (Murc) in Complex with Uma and Anp.
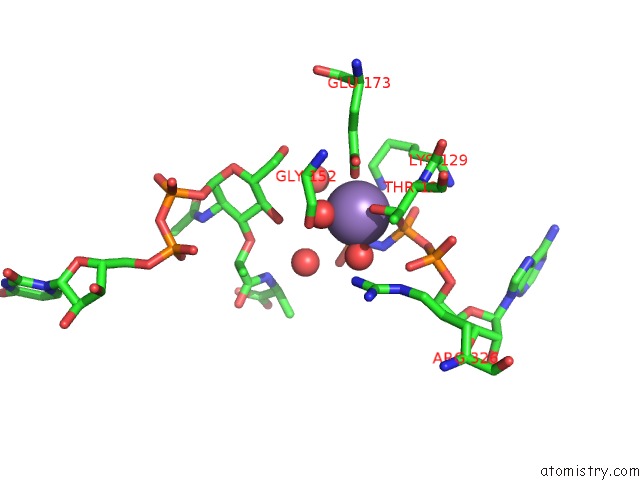
Mono view
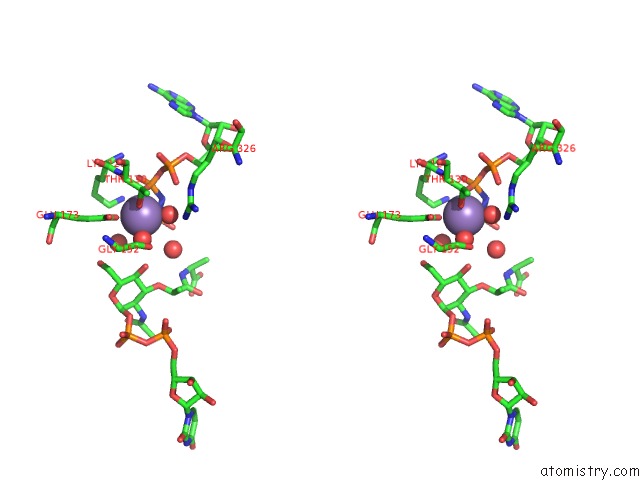
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Crystal Structure of Udp-N-Acetylmuramic Acid:L-Alanine Ligase (Murc) in Complex with Uma and Anp. within 5.0Å range:
|
Manganese binding site 3 out of 4 in 1p3d
Go back to
Manganese binding site 3 out
of 4 in the Crystal Structure of Udp-N-Acetylmuramic Acid:L-Alanine Ligase (Murc) in Complex with Uma and Anp.
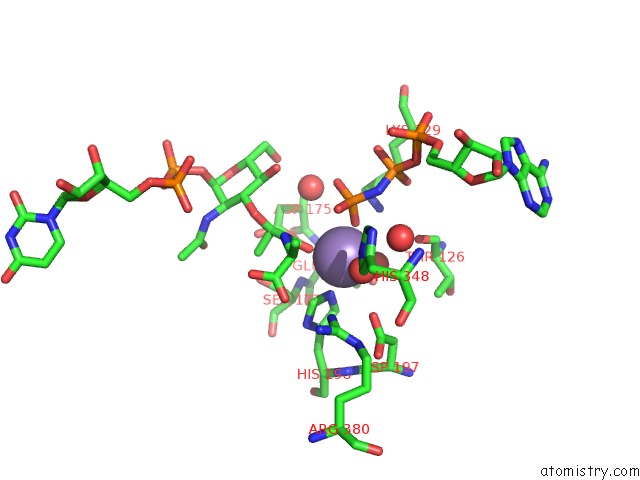
Mono view
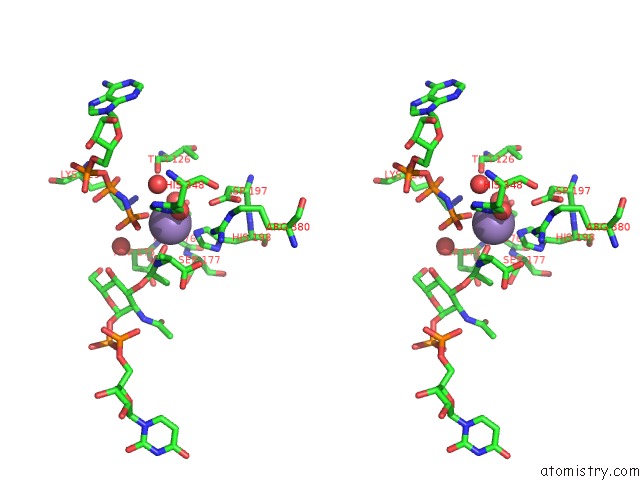
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Crystal Structure of Udp-N-Acetylmuramic Acid:L-Alanine Ligase (Murc) in Complex with Uma and Anp. within 5.0Å range:
|
Manganese binding site 4 out of 4 in 1p3d
Go back to
Manganese binding site 4 out
of 4 in the Crystal Structure of Udp-N-Acetylmuramic Acid:L-Alanine Ligase (Murc) in Complex with Uma and Anp.
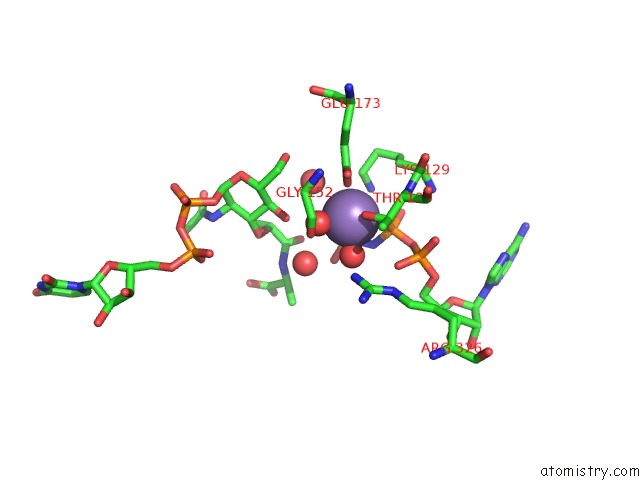
Mono view
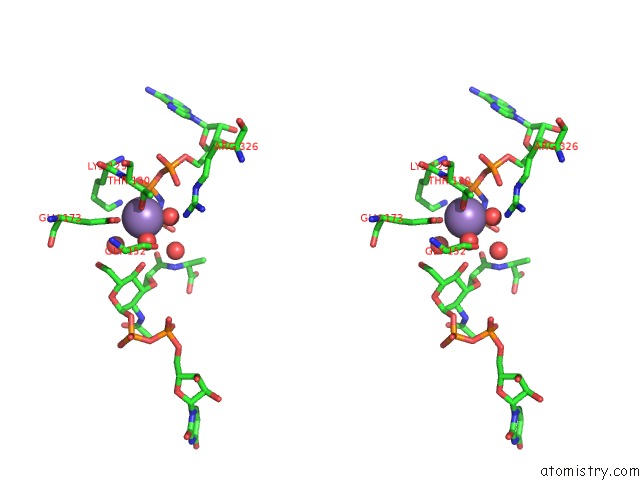
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of Crystal Structure of Udp-N-Acetylmuramic Acid:L-Alanine Ligase (Murc) in Complex with Uma and Anp. within 5.0Å range:
|
Reference:
C.D.Mol,
A.Brooun,
D.R.Dougan,
M.T.Hilgers,
L.W.Tari,
R.A.Wijnands,
M.W.Knuth,
D.E.Mcree,
R.V.Swanson.
Crystal Structures of Active Fully Assembled Substrate- and Product-Bound Complexes of Udp-N-Acetylmuramic Acid:L-Alanine Ligase (Murc) From Haemophilus Influenzae. J.Bacteriol. V. 185 4152 2003.
ISSN: ISSN 0021-9193
PubMed: 12837790
DOI: 10.1128/JB.185.14.4152-4162.2003
Page generated: Sat Oct 5 12:02:19 2024
ISSN: ISSN 0021-9193
PubMed: 12837790
DOI: 10.1128/JB.185.14.4152-4162.2003
Last articles
K in 3MLBK in 3MIO
K in 3MIJ
K in 3MD7
K in 3M62
K in 3M63
K in 3LNM
K in 3LUT
K in 3LIB
K in 3LQX