Manganese »
PDB 1n0n-1o99 »
1n51 »
Manganese in PDB 1n51: Aminopeptidase P in Complex with the Inhibitor Apstatin
Enzymatic activity of Aminopeptidase P in Complex with the Inhibitor Apstatin
All present enzymatic activity of Aminopeptidase P in Complex with the Inhibitor Apstatin:
3.4.11.9;
3.4.11.9;
Protein crystallography data
The structure of Aminopeptidase P in Complex with the Inhibitor Apstatin, PDB code: 1n51
was solved by
S.C.Graham,
M.J.Maher,
M.H.Lee,
W.H.Simmons,
H.C.Freeman,
J.M.Guss,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.90 / 2.30 |
| Space group | I 41 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 139.319, 139.319, 231.005, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.9 / 20.4 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Aminopeptidase P in Complex with the Inhibitor Apstatin
(pdb code 1n51). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 3 binding sites of Manganese where determined in the Aminopeptidase P in Complex with the Inhibitor Apstatin, PDB code: 1n51:
Jump to Manganese binding site number: 1; 2; 3;
In total 3 binding sites of Manganese where determined in the Aminopeptidase P in Complex with the Inhibitor Apstatin, PDB code: 1n51:
Jump to Manganese binding site number: 1; 2; 3;
Manganese binding site 1 out of 3 in 1n51
Go back to
Manganese binding site 1 out
of 3 in the Aminopeptidase P in Complex with the Inhibitor Apstatin
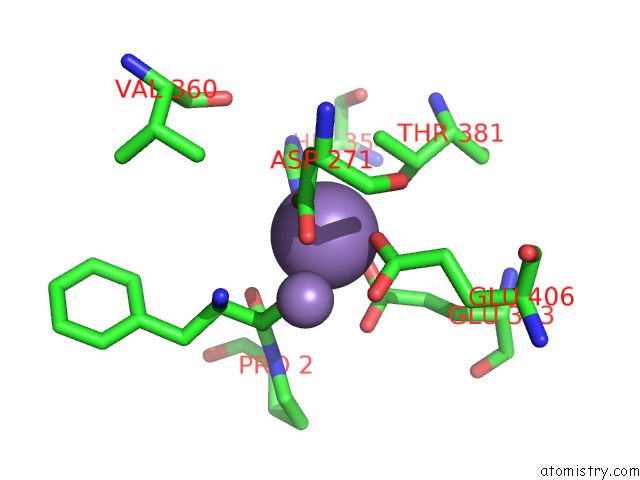
Mono view
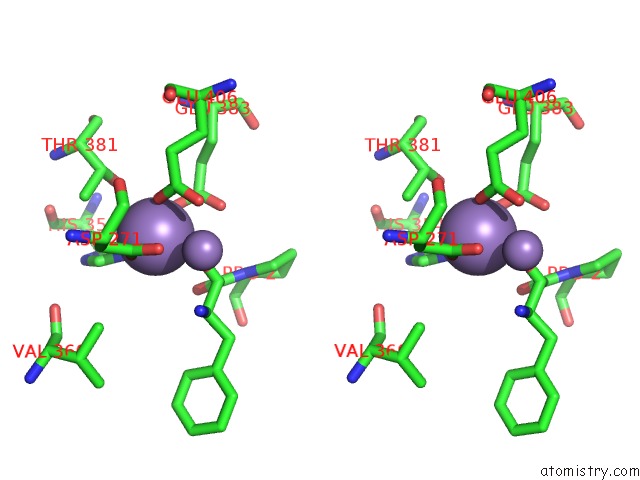
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Aminopeptidase P in Complex with the Inhibitor Apstatin within 5.0Å range:
|
Manganese binding site 2 out of 3 in 1n51
Go back to
Manganese binding site 2 out
of 3 in the Aminopeptidase P in Complex with the Inhibitor Apstatin
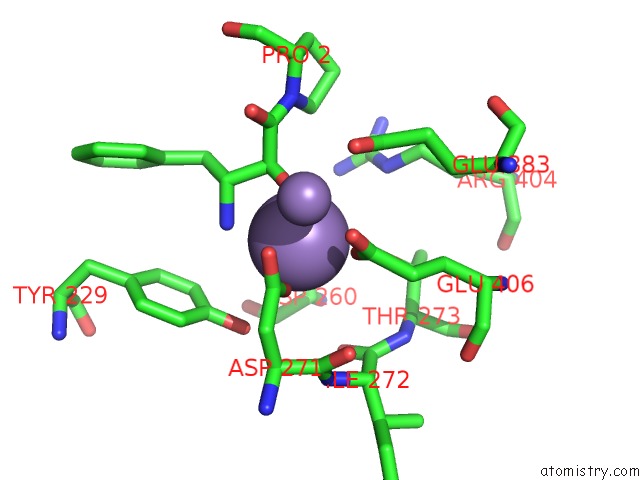
Mono view
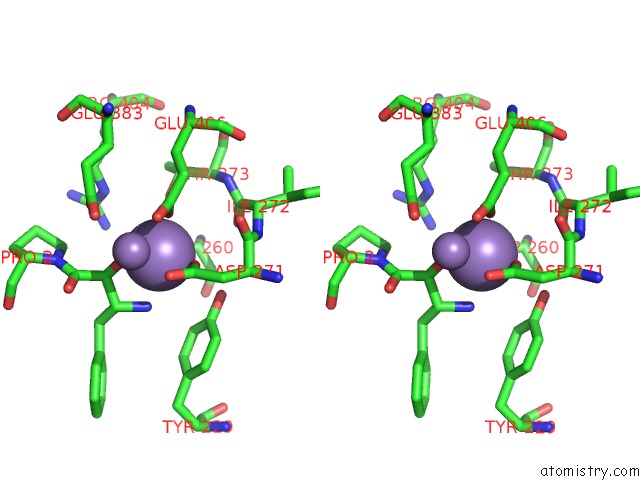
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Aminopeptidase P in Complex with the Inhibitor Apstatin within 5.0Å range:
|
Manganese binding site 3 out of 3 in 1n51
Go back to
Manganese binding site 3 out
of 3 in the Aminopeptidase P in Complex with the Inhibitor Apstatin
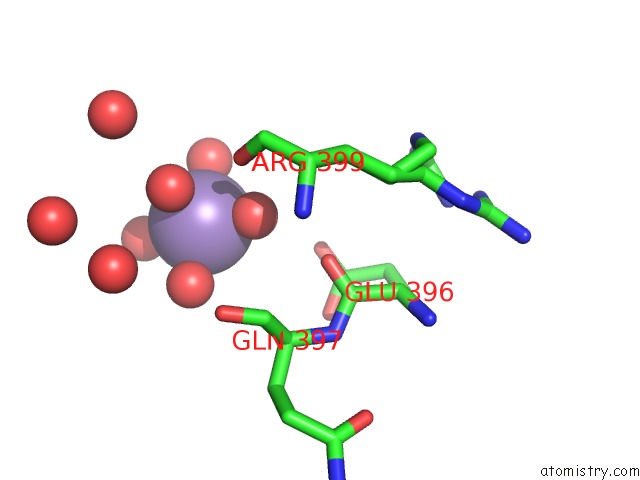
Mono view
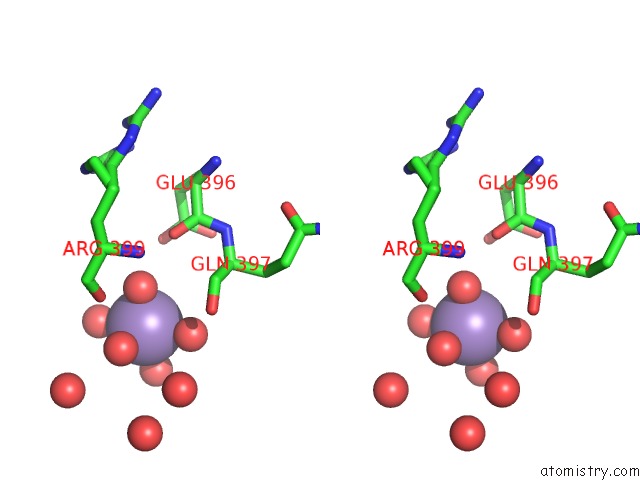
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Aminopeptidase P in Complex with the Inhibitor Apstatin within 5.0Å range:
|
Reference:
S.C.Graham,
M.J.Maher,
W.H.Simmons,
H.C.Freeman,
J.M.Guss.
Structure of Escherichia Coli Aminopeptidase P in Complex with the Inhibitor Apstatin. Acta Crystallogr.,Sect.D V. 60 1770 2004.
ISSN: ISSN 0907-4449
PubMed: 15388923
DOI: 10.1107/S0907444904018724
Page generated: Sat Oct 5 11:50:08 2024
ISSN: ISSN 0907-4449
PubMed: 15388923
DOI: 10.1107/S0907444904018724
Last articles
K in 8OLVK in 8OLS
K in 8OLW
K in 8OLJ
K in 8OFD
K in 8OEO
K in 8OED
K in 8OEH
K in 8K1Z
K in 8K1U