Manganese »
PDB 8wq7-9c4d »
9asc »
Manganese in PDB 9asc: Cryo-Em Structure of the Glycosyltransferase Arnc From Salmonella Enterica in the Udp-Bound State Determined on Talos Arctica Microscope
Enzymatic activity of Cryo-Em Structure of the Glycosyltransferase Arnc From Salmonella Enterica in the Udp-Bound State Determined on Talos Arctica Microscope
All present enzymatic activity of Cryo-Em Structure of the Glycosyltransferase Arnc From Salmonella Enterica in the Udp-Bound State Determined on Talos Arctica Microscope:
2.4.2.53;
2.4.2.53;
Manganese Binding Sites:
The binding sites of Manganese atom in the Cryo-Em Structure of the Glycosyltransferase Arnc From Salmonella Enterica in the Udp-Bound State Determined on Talos Arctica Microscope
(pdb code 9asc). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 4 binding sites of Manganese where determined in the Cryo-Em Structure of the Glycosyltransferase Arnc From Salmonella Enterica in the Udp-Bound State Determined on Talos Arctica Microscope, PDB code: 9asc:
Jump to Manganese binding site number: 1; 2; 3; 4;
In total 4 binding sites of Manganese where determined in the Cryo-Em Structure of the Glycosyltransferase Arnc From Salmonella Enterica in the Udp-Bound State Determined on Talos Arctica Microscope, PDB code: 9asc:
Jump to Manganese binding site number: 1; 2; 3; 4;
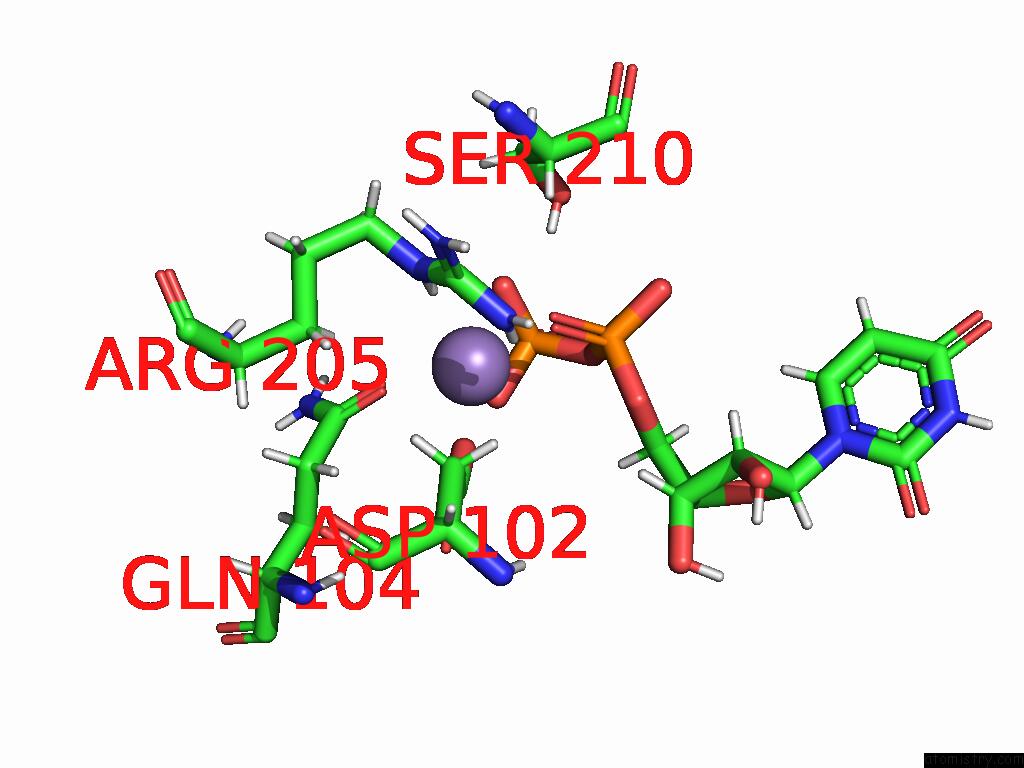
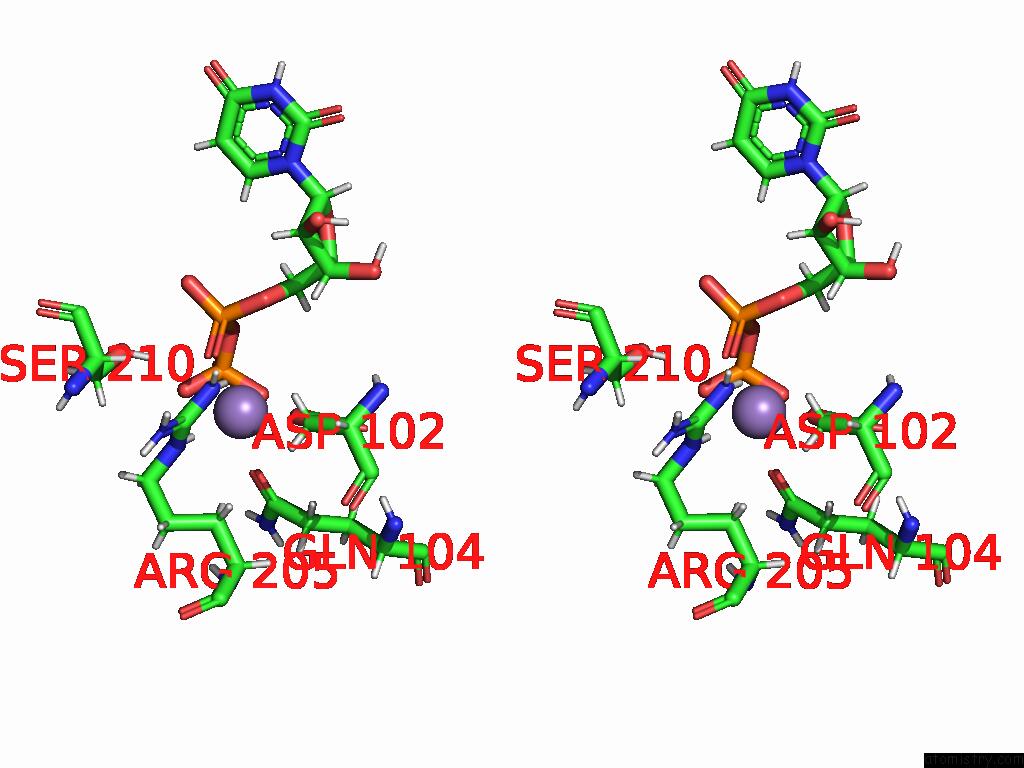
Manganese binding site 1 out of 4 in 9asc
Go back to
Manganese binding site 1 out
of 4 in the Cryo-Em Structure of the Glycosyltransferase Arnc From Salmonella Enterica in the Udp-Bound State Determined on Talos Arctica Microscope
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Cryo-Em Structure of the Glycosyltransferase Arnc From Salmonella Enterica in the Udp-Bound State Determined on Talos Arctica Microscope within 5.0Å range:
|
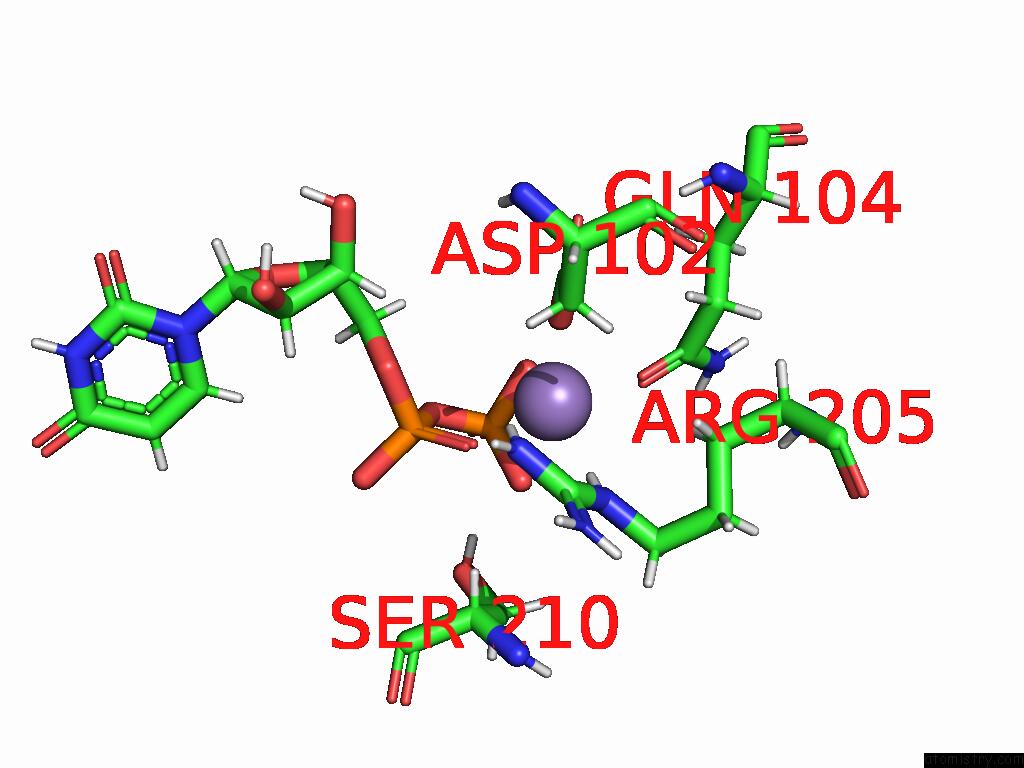
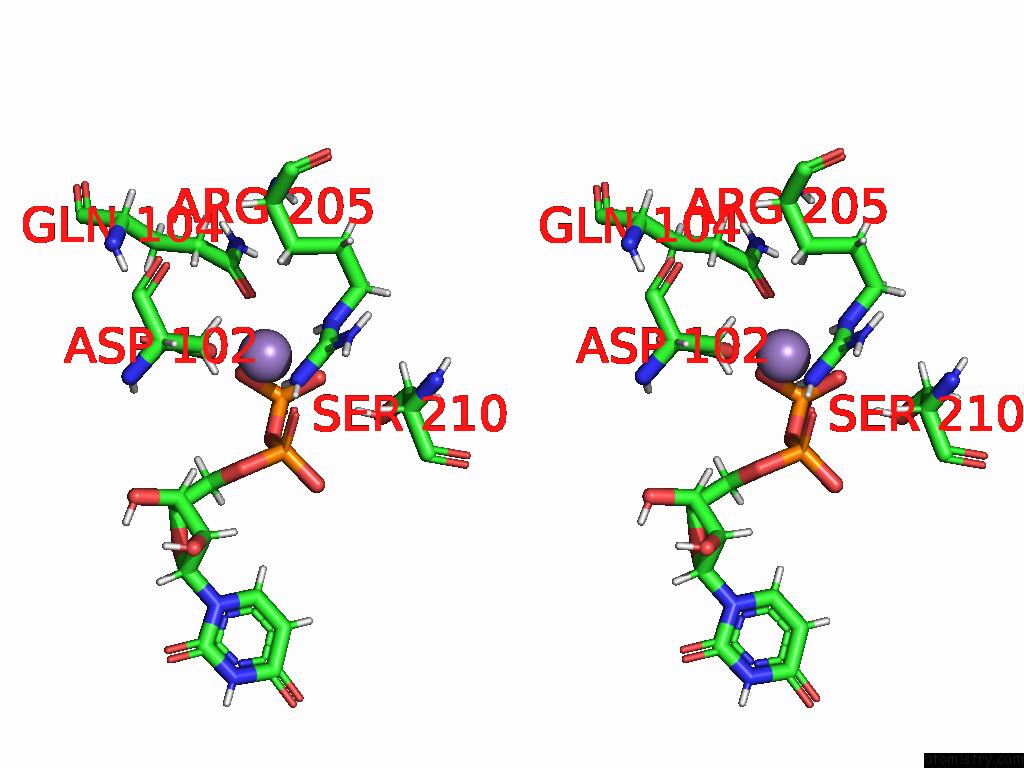
Manganese binding site 2 out of 4 in 9asc
Go back to
Manganese binding site 2 out
of 4 in the Cryo-Em Structure of the Glycosyltransferase Arnc From Salmonella Enterica in the Udp-Bound State Determined on Talos Arctica Microscope
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Cryo-Em Structure of the Glycosyltransferase Arnc From Salmonella Enterica in the Udp-Bound State Determined on Talos Arctica Microscope within 5.0Å range:
|
Manganese binding site 3 out of 4 in 9asc
Go back to
Manganese binding site 3 out
of 4 in the Cryo-Em Structure of the Glycosyltransferase Arnc From Salmonella Enterica in the Udp-Bound State Determined on Talos Arctica Microscope
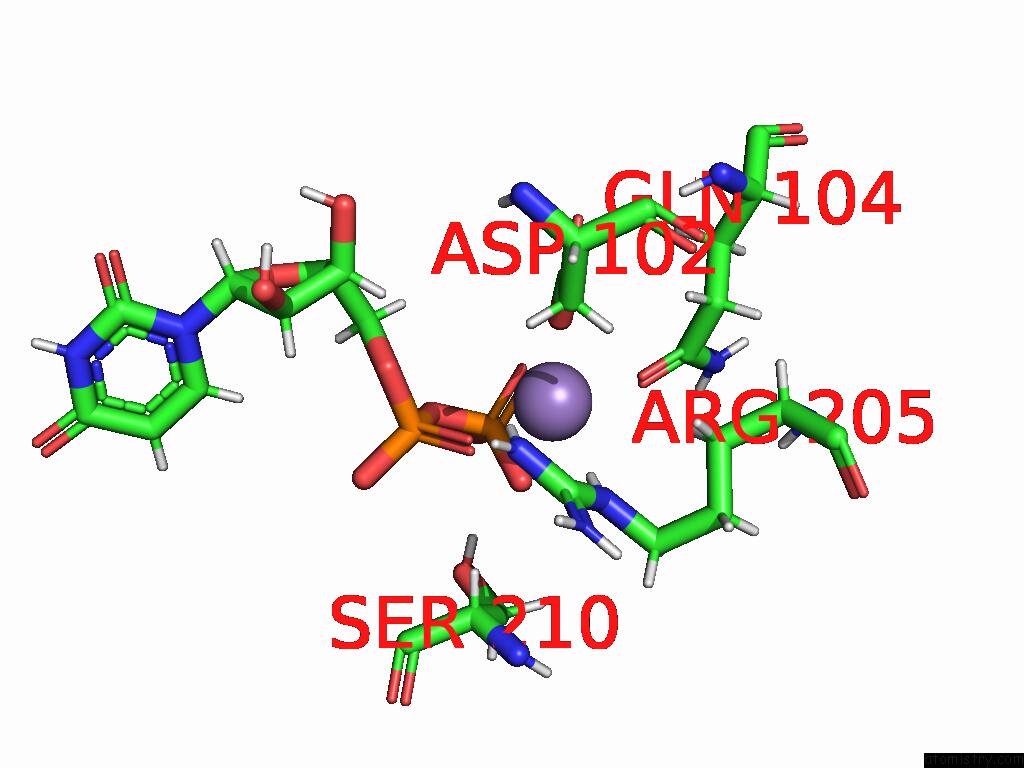
Mono view
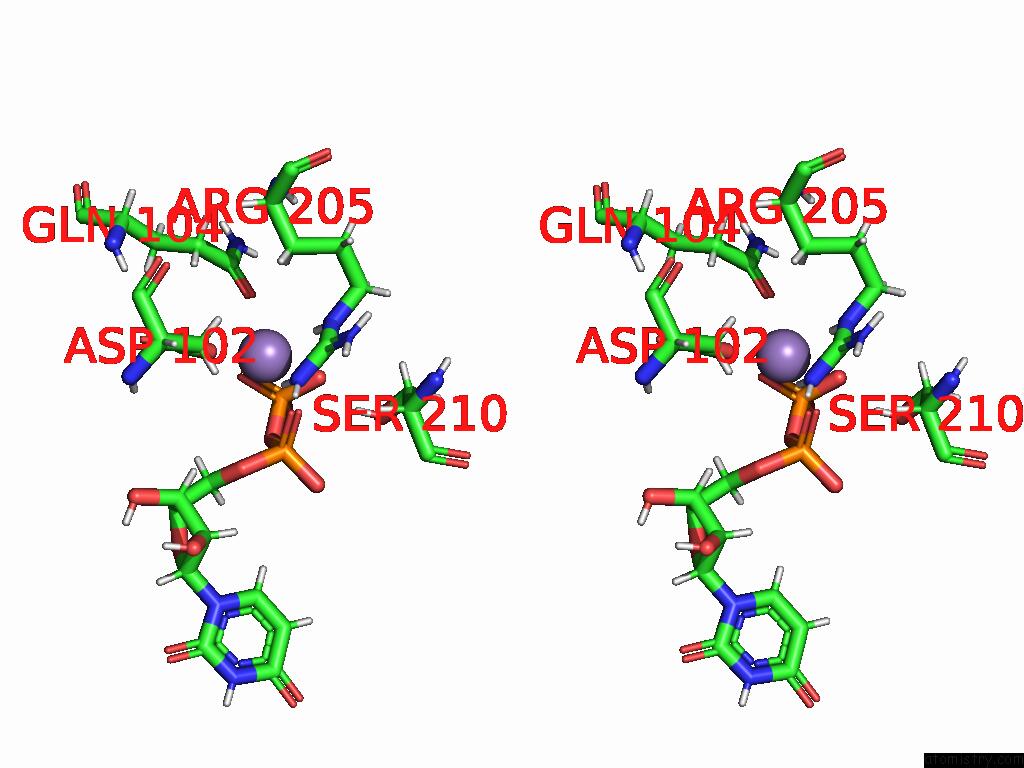
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Cryo-Em Structure of the Glycosyltransferase Arnc From Salmonella Enterica in the Udp-Bound State Determined on Talos Arctica Microscope within 5.0Å range:
|
Manganese binding site 4 out of 4 in 9asc
Go back to
Manganese binding site 4 out
of 4 in the Cryo-Em Structure of the Glycosyltransferase Arnc From Salmonella Enterica in the Udp-Bound State Determined on Talos Arctica Microscope
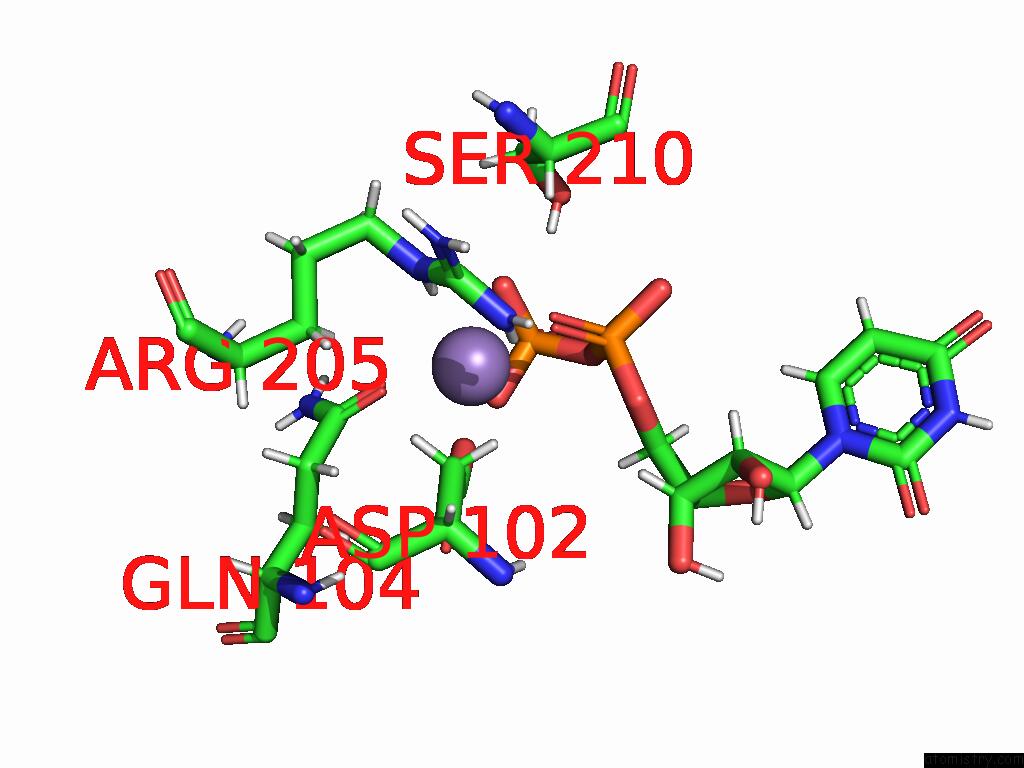
Mono view
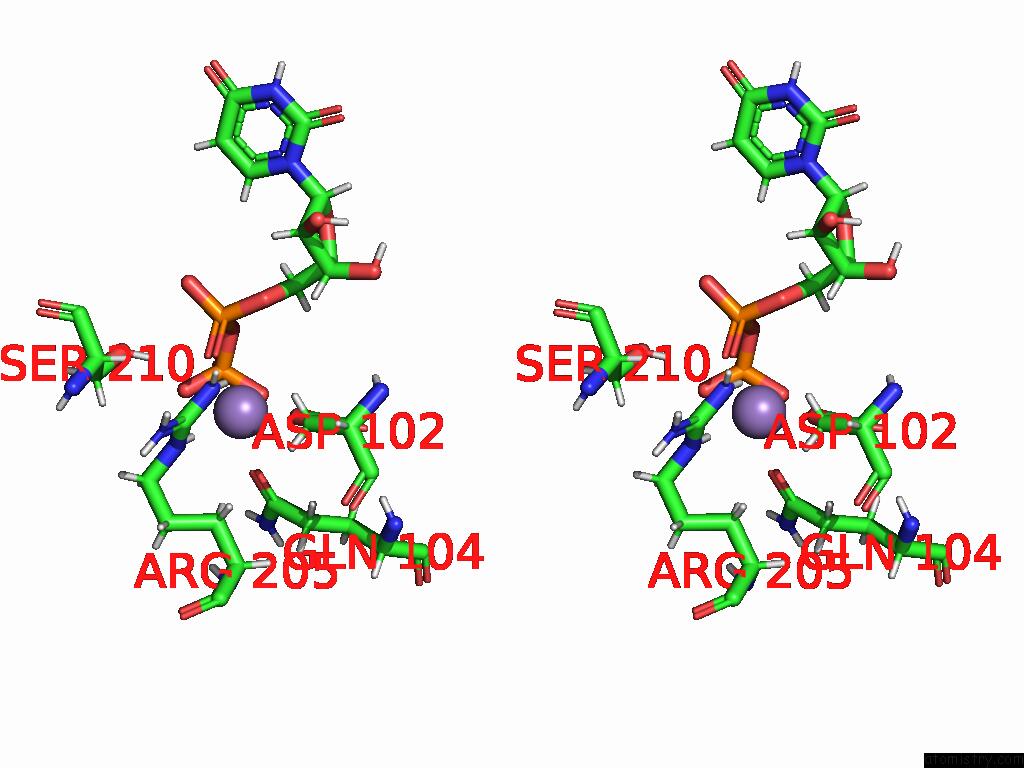
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of Cryo-Em Structure of the Glycosyltransferase Arnc From Salmonella Enterica in the Udp-Bound State Determined on Talos Arctica Microscope within 5.0Å range:
|
Reference:
K.U.Ashraf,
M.Bunoro-Batista,
T.B.Ansell,
A.Punetha,
S.Rosario-Garrido,
E.Firlar,
J.T.Kaelber,
P.J.Stansfeld,
V.I.Petrou.
Structure and Mechanism of the Polyprenyl Phosphate Glycosyltransferase Arnc Participating in Aminoarabinose Biosynthesis in Gram-Negative Bacteria To Be Published.
Page generated: Sun Feb 9 08:27:47 2025
Last articles
Zn in 9MJ5Zn in 9HNW
Zn in 9G0L
Zn in 9FNE
Zn in 9DZN
Zn in 9E0I
Zn in 9D32
Zn in 9DAK
Zn in 8ZXC
Zn in 8ZUF