Manganese »
PDB 8sln-8uwb »
8umc »
Manganese in PDB 8umc: Deinococcus Aerius TR0125 C-Glucosyl Deglycosidase (Cgd), Cryo-Em
Manganese Binding Sites:
The binding sites of Manganese atom in the Deinococcus Aerius TR0125 C-Glucosyl Deglycosidase (Cgd), Cryo-Em
(pdb code 8umc). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 2 binding sites of Manganese where determined in the Deinococcus Aerius TR0125 C-Glucosyl Deglycosidase (Cgd), Cryo-Em, PDB code: 8umc:
Jump to Manganese binding site number: 1; 2;
In total 2 binding sites of Manganese where determined in the Deinococcus Aerius TR0125 C-Glucosyl Deglycosidase (Cgd), Cryo-Em, PDB code: 8umc:
Jump to Manganese binding site number: 1; 2;
Manganese binding site 1 out of 2 in 8umc
Go back to
Manganese binding site 1 out
of 2 in the Deinococcus Aerius TR0125 C-Glucosyl Deglycosidase (Cgd), Cryo-Em
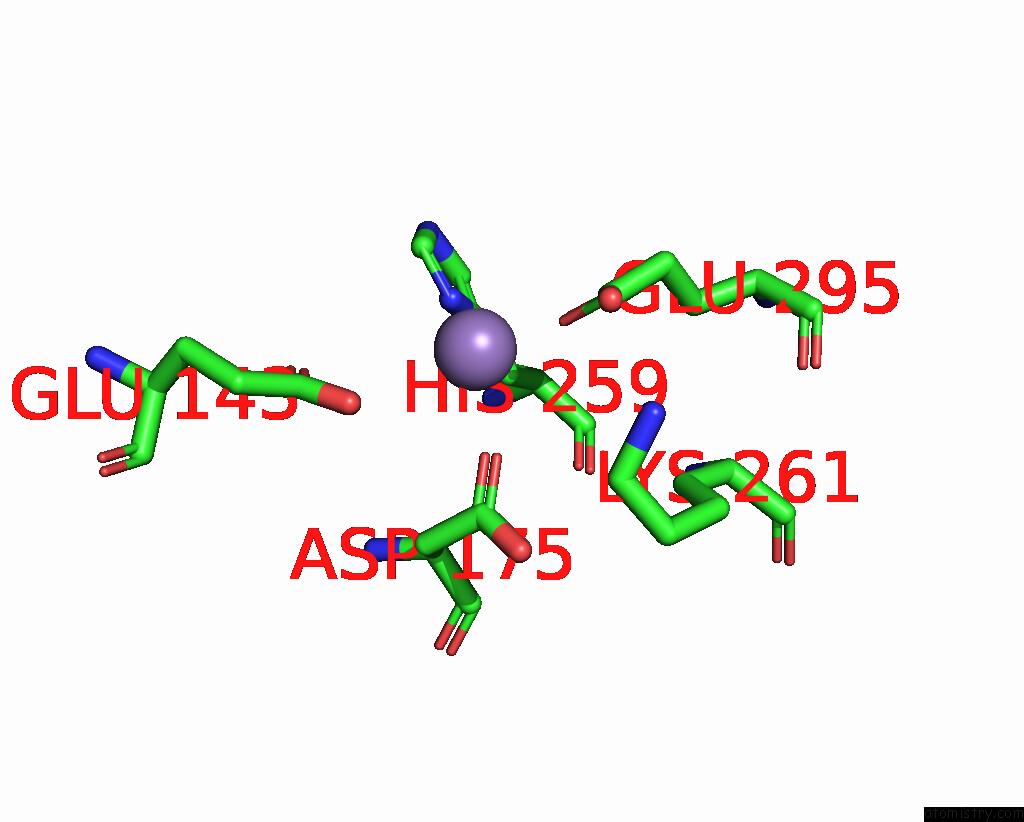
Mono view
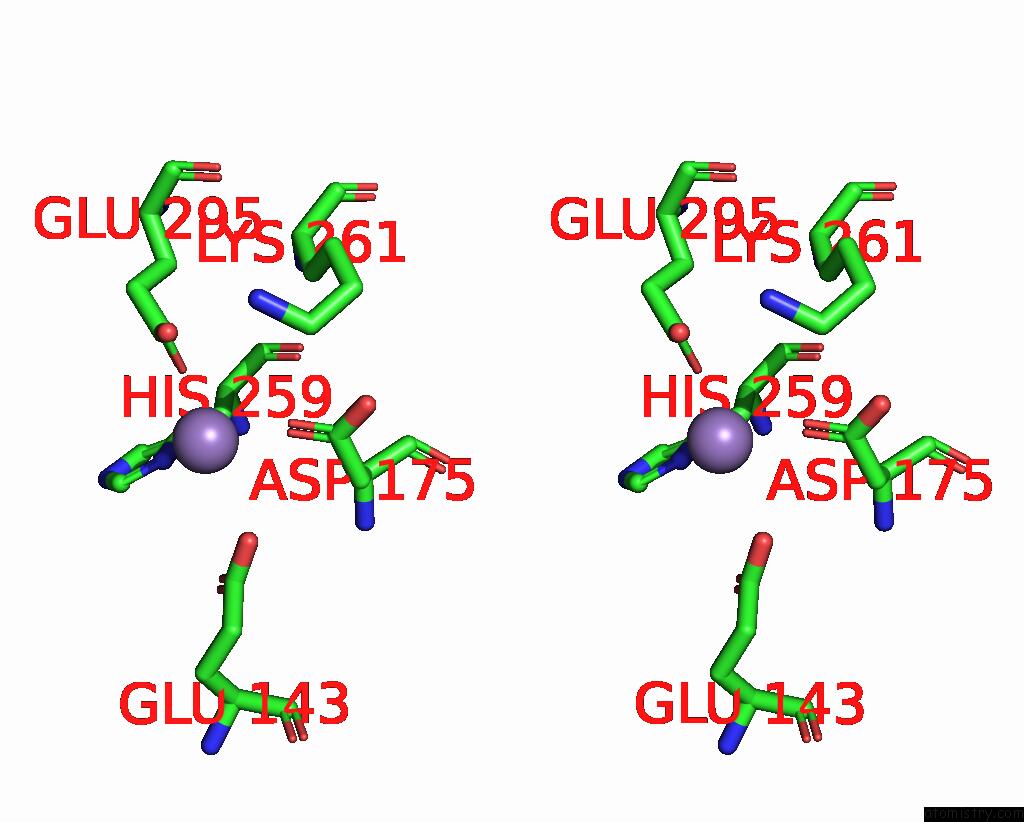
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Deinococcus Aerius TR0125 C-Glucosyl Deglycosidase (Cgd), Cryo-Em within 5.0Å range:
|
Manganese binding site 2 out of 2 in 8umc
Go back to
Manganese binding site 2 out
of 2 in the Deinococcus Aerius TR0125 C-Glucosyl Deglycosidase (Cgd), Cryo-Em
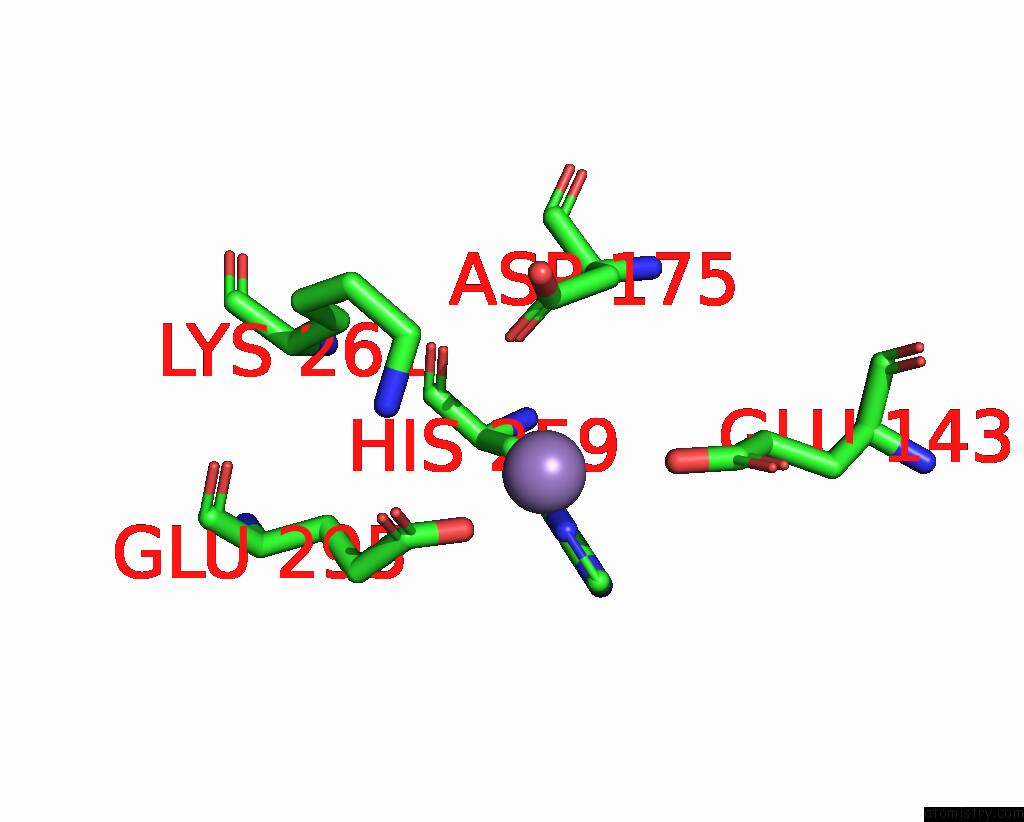
Mono view
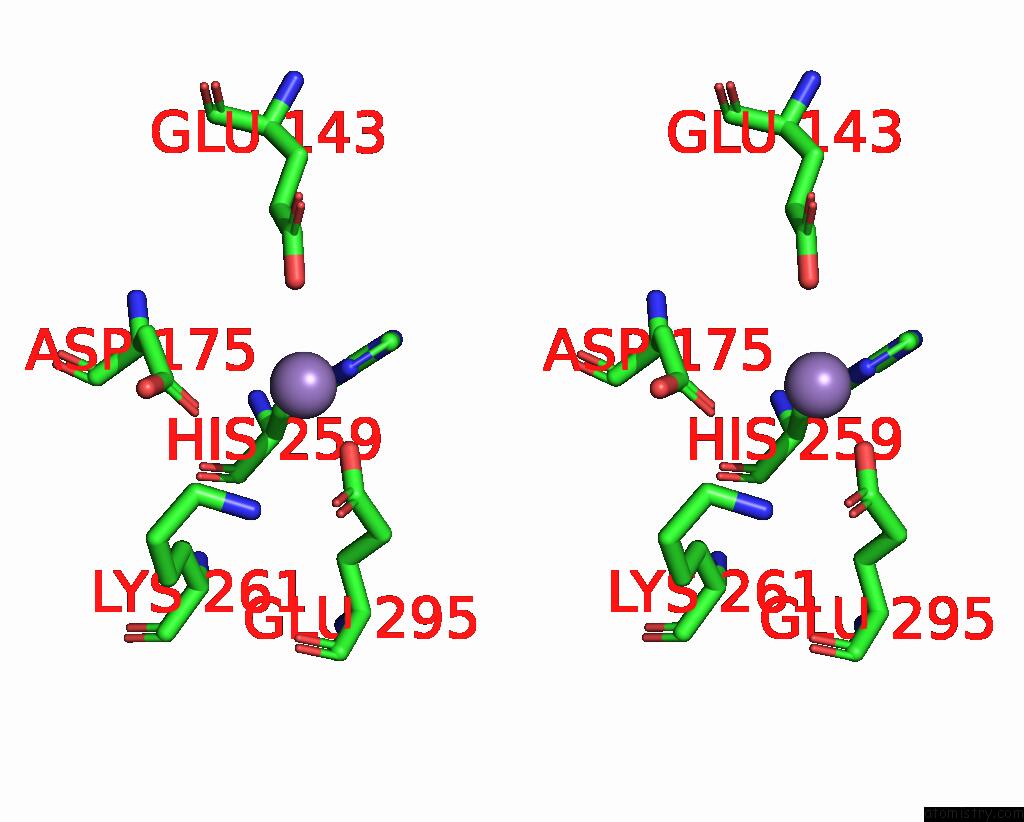
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Deinococcus Aerius TR0125 C-Glucosyl Deglycosidase (Cgd), Cryo-Em within 5.0Å range:
|
Reference:
V.Furlanetto,
D.C.Kalyani,
A.Kostelac,
J.Puc,
D.Haltrich,
B.M.Hallberg,
C.Divne.
Structural and Functional Characterization of A Gene Cluster Responsible For Deglycosylation of C-Glucosyl Flavonoids and Xanthonoids By Deinococcus Aerius. J.Mol.Biol. V. 436 68547 2024.
ISSN: ESSN 1089-8638
PubMed: 38508304
DOI: 10.1016/J.JMB.2024.168547
Page generated: Sun Oct 6 14:01:28 2024
ISSN: ESSN 1089-8638
PubMed: 38508304
DOI: 10.1016/J.JMB.2024.168547
Last articles
Zn in 9MJ5Zn in 9HNW
Zn in 9G0L
Zn in 9FNE
Zn in 9DZN
Zn in 9E0I
Zn in 9D32
Zn in 9DAK
Zn in 8ZXC
Zn in 8ZUF