Manganese »
PDB 7x9j-7yzp »
7yzi »
Manganese in PDB 7yzi: Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya
Enzymatic activity of Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya
All present enzymatic activity of Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya:
4.6.1.1;
4.6.1.1;
Manganese Binding Sites:
The binding sites of Manganese atom in the Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya
(pdb code 7yzi). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 4 binding sites of Manganese where determined in the Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya, PDB code: 7yzi:
Jump to Manganese binding site number: 1; 2; 3; 4;
In total 4 binding sites of Manganese where determined in the Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya, PDB code: 7yzi:
Jump to Manganese binding site number: 1; 2; 3; 4;
Manganese binding site 1 out of 4 in 7yzi
Go back to
Manganese binding site 1 out
of 4 in the Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya
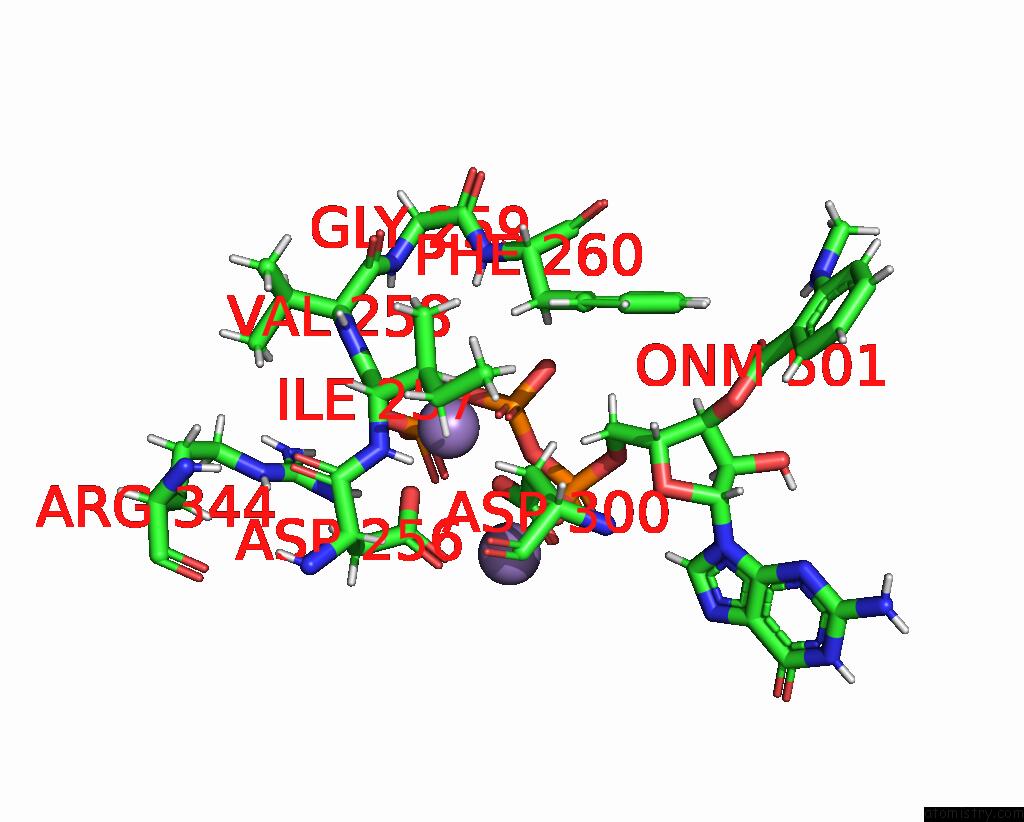
Mono view
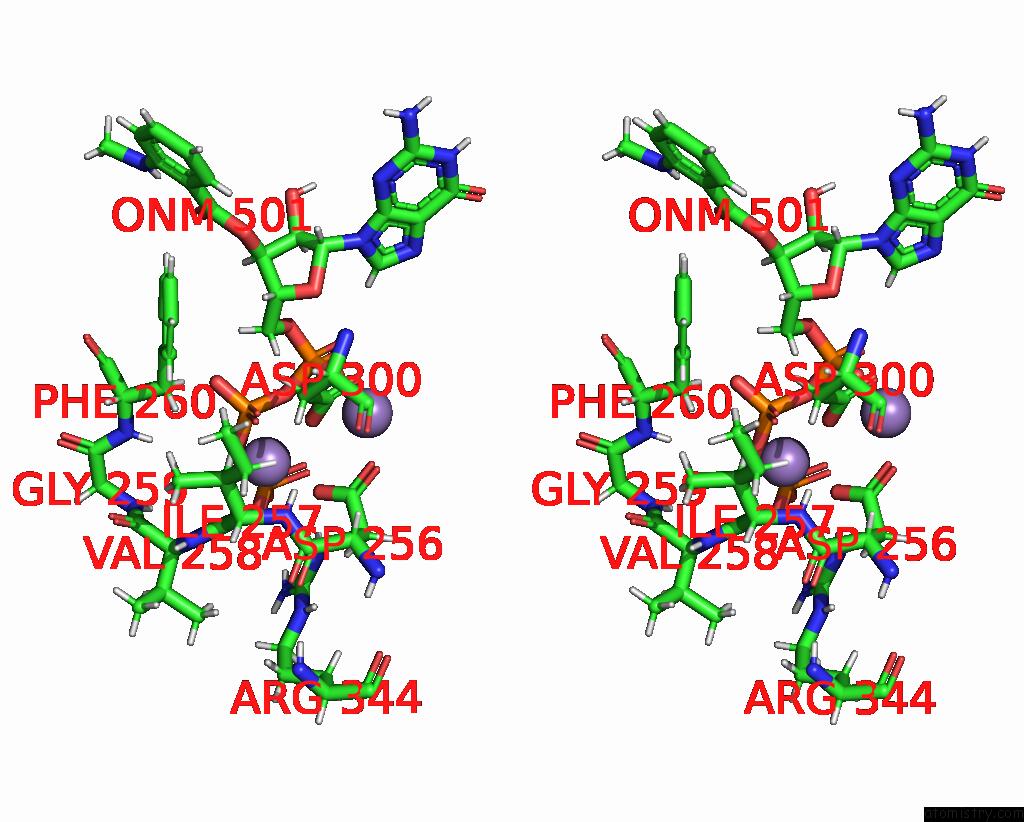
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya within 5.0Å range:
|
Manganese binding site 2 out of 4 in 7yzi
Go back to
Manganese binding site 2 out
of 4 in the Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya
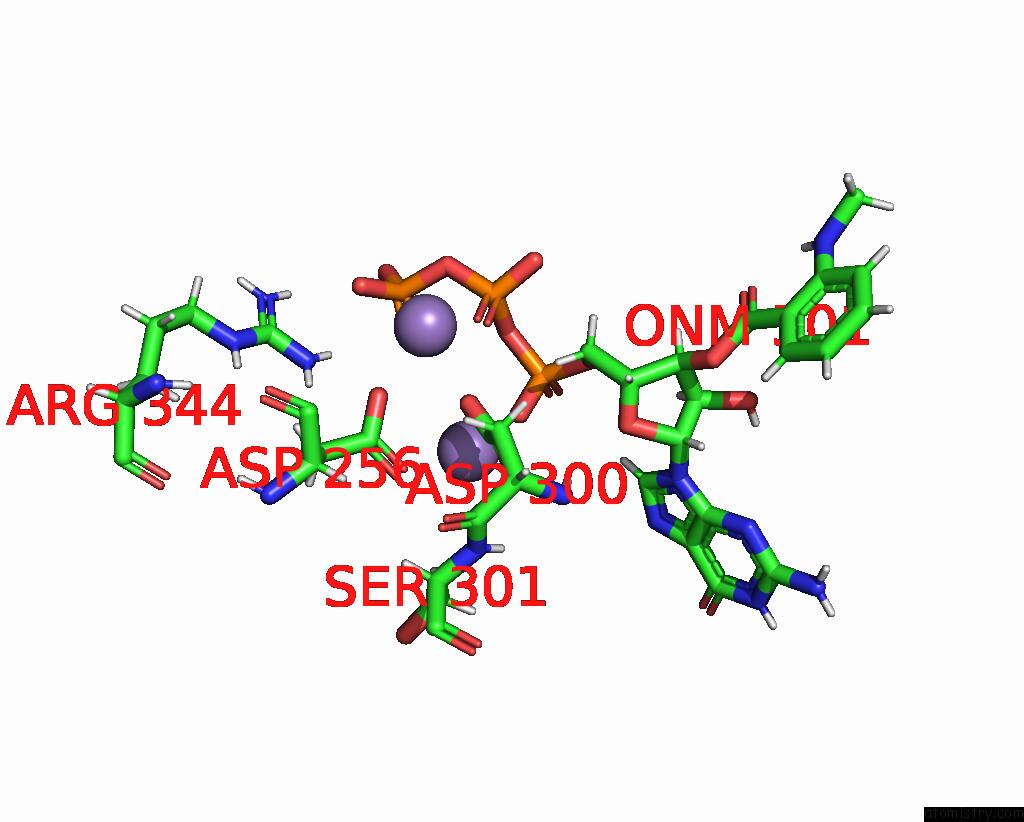
Mono view
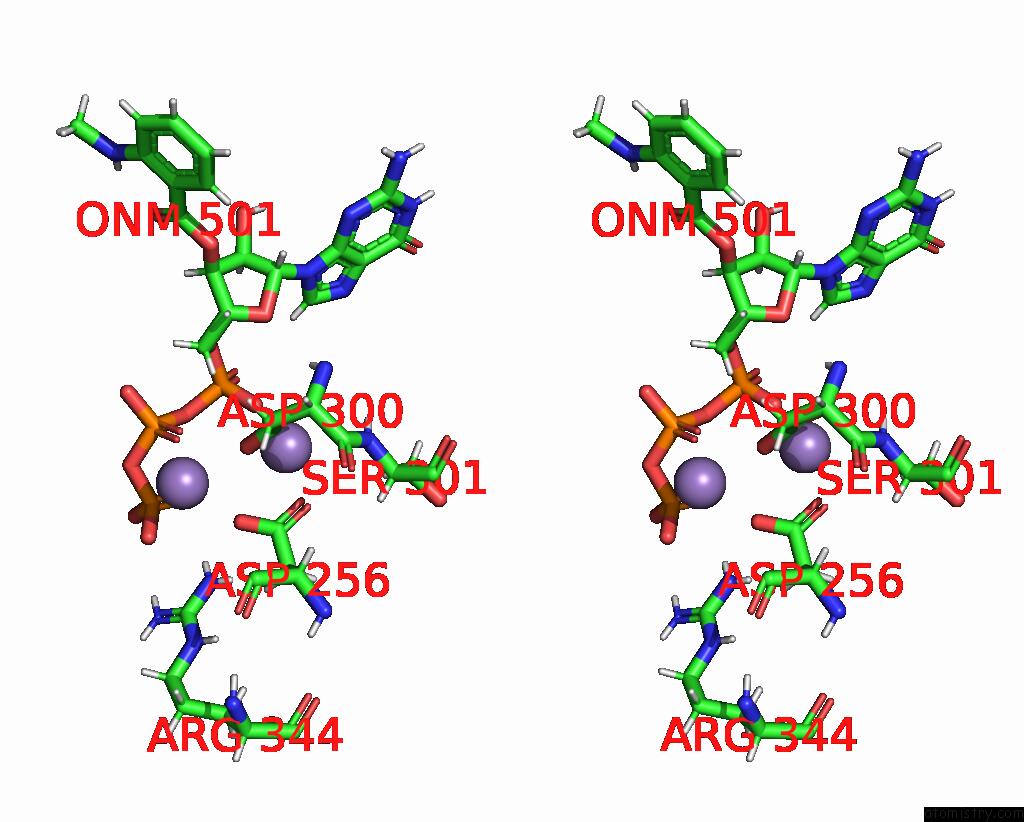
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya within 5.0Å range:
|
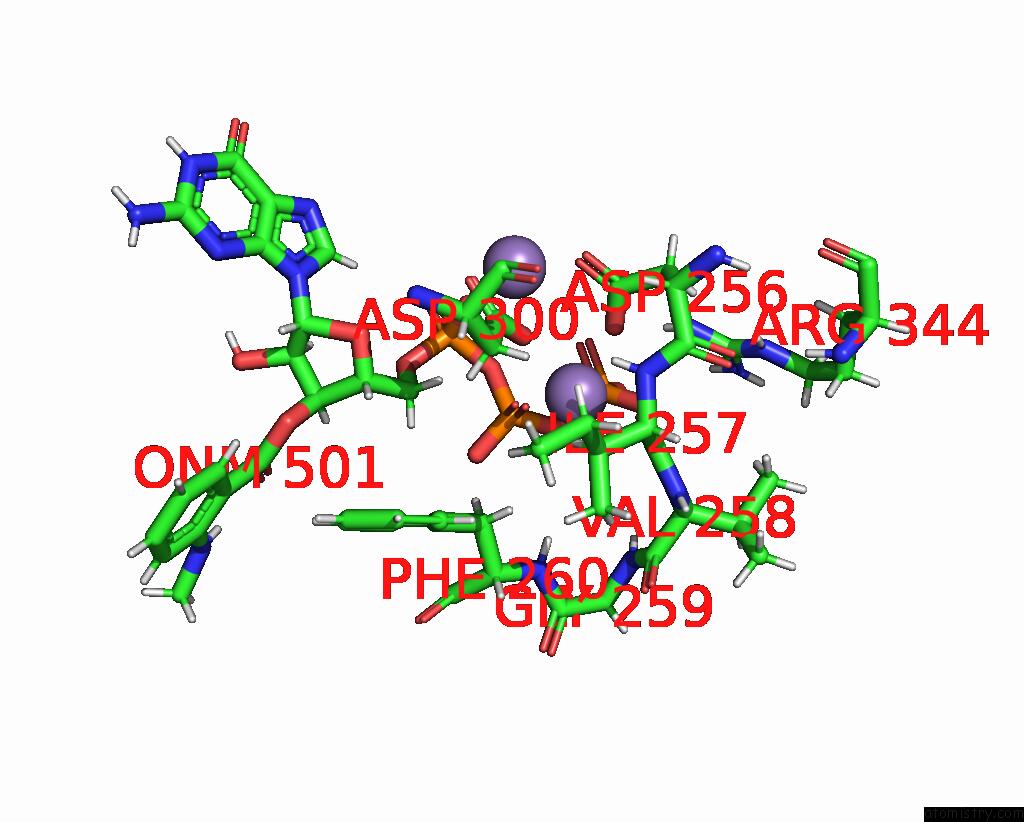
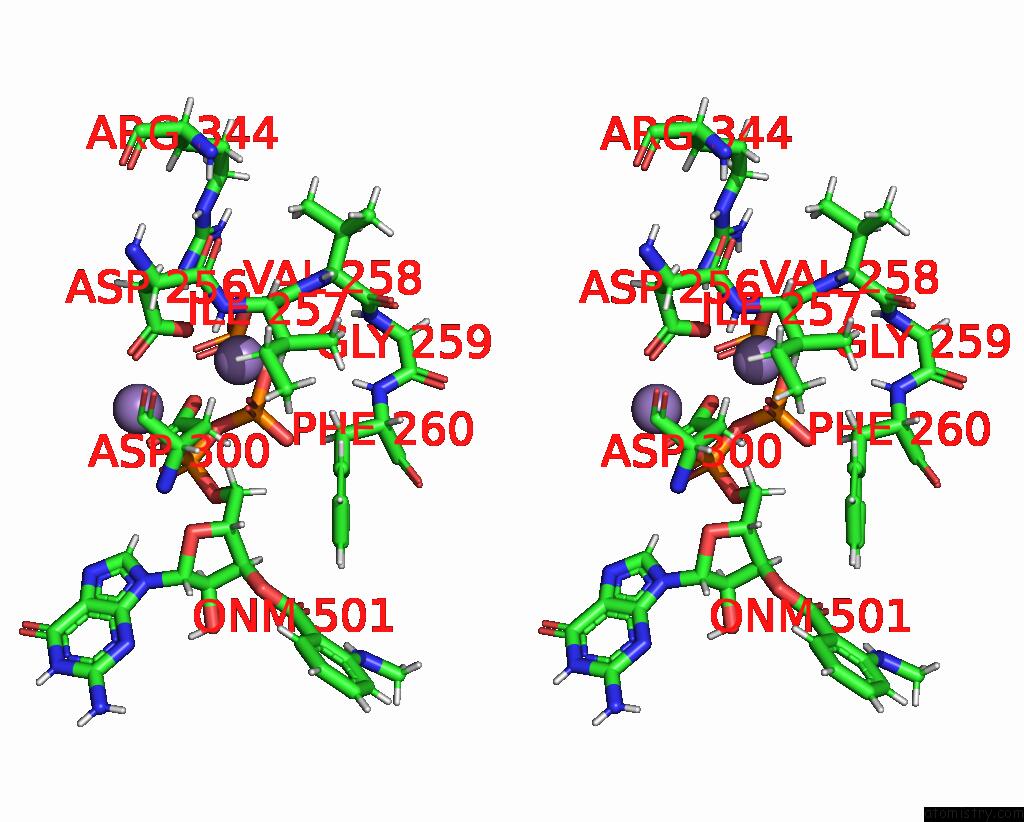
Manganese binding site 3 out of 4 in 7yzi
Go back to
Manganese binding site 3 out
of 4 in the Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya within 5.0Å range:
|
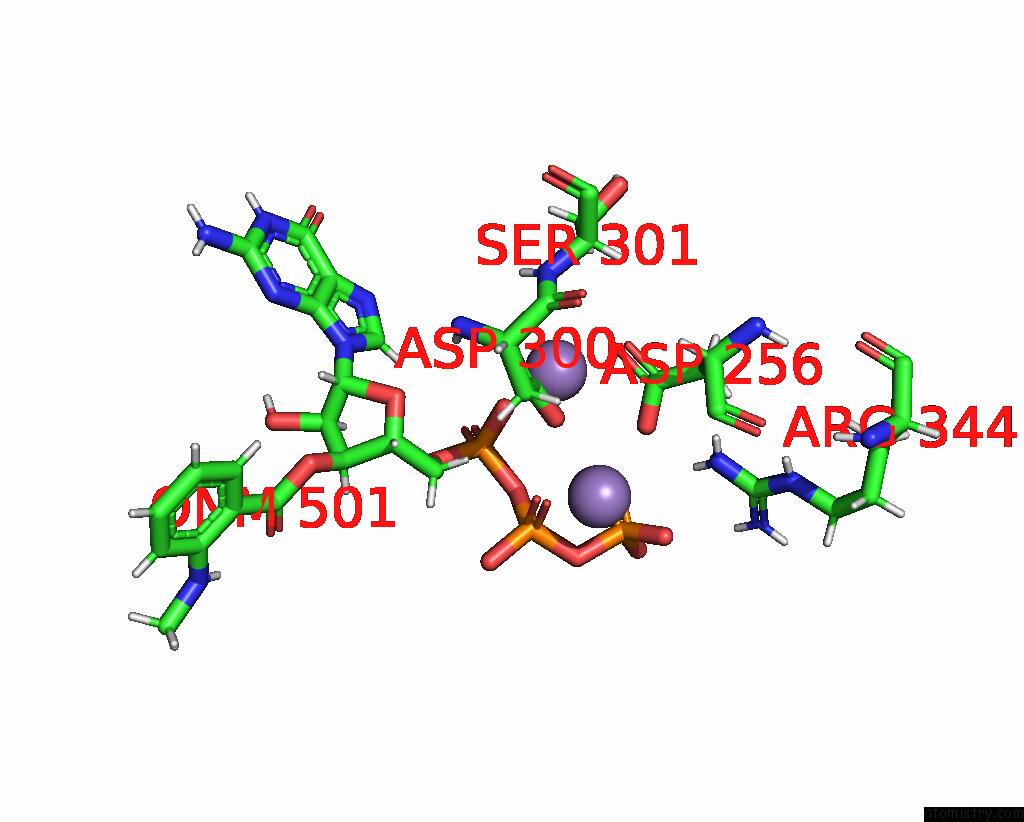
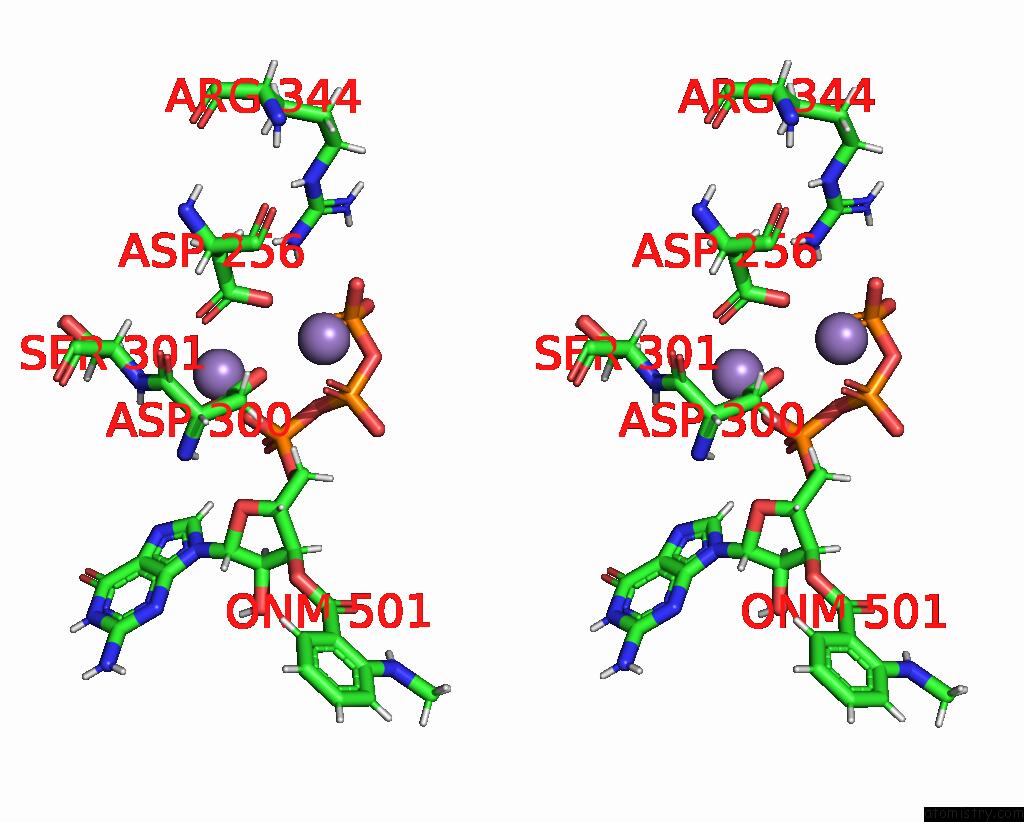
Manganese binding site 4 out of 4 in 7yzi
Go back to
Manganese binding site 4 out
of 4 in the Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of Structure of Mycobacterium Tuberculosis Adenylyl Cyclase RV1625C / Cya within 5.0Å range:
|
Reference:
V.Mehta,
B.Khanppnavar,
D.Schuster,
I.Kantarci,
I.Vercellino,
A.Kosturanova,
T.Iype,
S.Stefanic,
P.Picotti,
V.M.Korkhov.
Structure of Mycobacterium Tuberculosis Cya, An Evolutionary Ancestor of the Mammalian Membrane Adenylyl Cyclases. Elife V. 11 2022.
ISSN: ESSN 2050-084X
PubMed: 35980026
DOI: 10.7554/ELIFE.77032
Page generated: Sun Oct 6 11:11:02 2024
ISSN: ESSN 2050-084X
PubMed: 35980026
DOI: 10.7554/ELIFE.77032
Last articles
F in 5FD2F in 5FET
F in 5FDP
F in 5FED
F in 5F9F
F in 5FCT
F in 5FC4
F in 5FCK
F in 5FBN
F in 5FBO