Manganese »
PDB 7uuh-7x9i »
7w7a »
Manganese in PDB 7w7a: Heme Exporter in Complex with Mn-Containing Protoporphyrin IX, Mn- Anomalous Data
Protein crystallography data
The structure of Heme Exporter in Complex with Mn-Containing Protoporphyrin IX, Mn- Anomalous Data, PDB code: 7w7a
was solved by
T.Hisano,
H.Nakamura,
M.M.Rahman,
T.Tosha,
M.Shirouzu,
Y.Shiro,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.82 / 3.20 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 82.488, 133.672, 159.838, 111.81, 99.83, 94.43 |
| R / Rfree (%) | 26.4 / 30.3 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Heme Exporter in Complex with Mn-Containing Protoporphyrin IX, Mn- Anomalous Data
(pdb code 7w7a). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 2 binding sites of Manganese where determined in the Heme Exporter in Complex with Mn-Containing Protoporphyrin IX, Mn- Anomalous Data, PDB code: 7w7a:
Jump to Manganese binding site number: 1; 2;
In total 2 binding sites of Manganese where determined in the Heme Exporter in Complex with Mn-Containing Protoporphyrin IX, Mn- Anomalous Data, PDB code: 7w7a:
Jump to Manganese binding site number: 1; 2;
Manganese binding site 1 out of 2 in 7w7a
Go back to
Manganese binding site 1 out
of 2 in the Heme Exporter in Complex with Mn-Containing Protoporphyrin IX, Mn- Anomalous Data
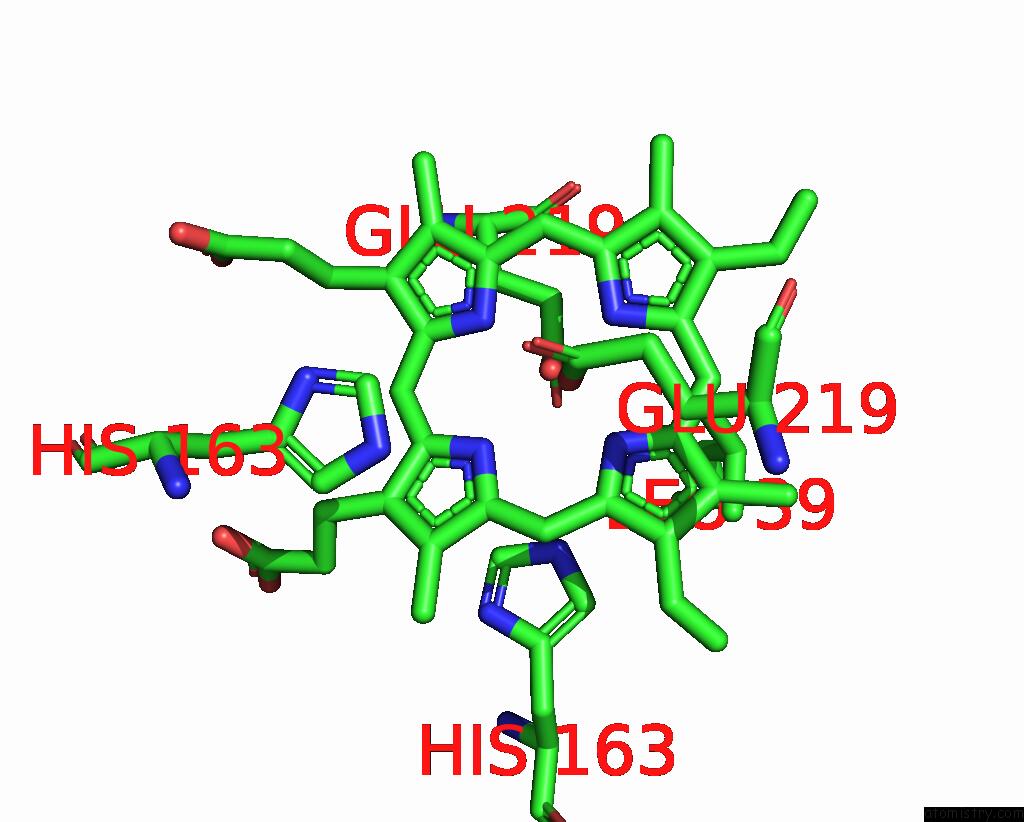
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Heme Exporter in Complex with Mn-Containing Protoporphyrin IX, Mn- Anomalous Data within 5.0Å range:
|
Manganese binding site 2 out of 2 in 7w7a
Go back to
Manganese binding site 2 out
of 2 in the Heme Exporter in Complex with Mn-Containing Protoporphyrin IX, Mn- Anomalous Data

Mono view
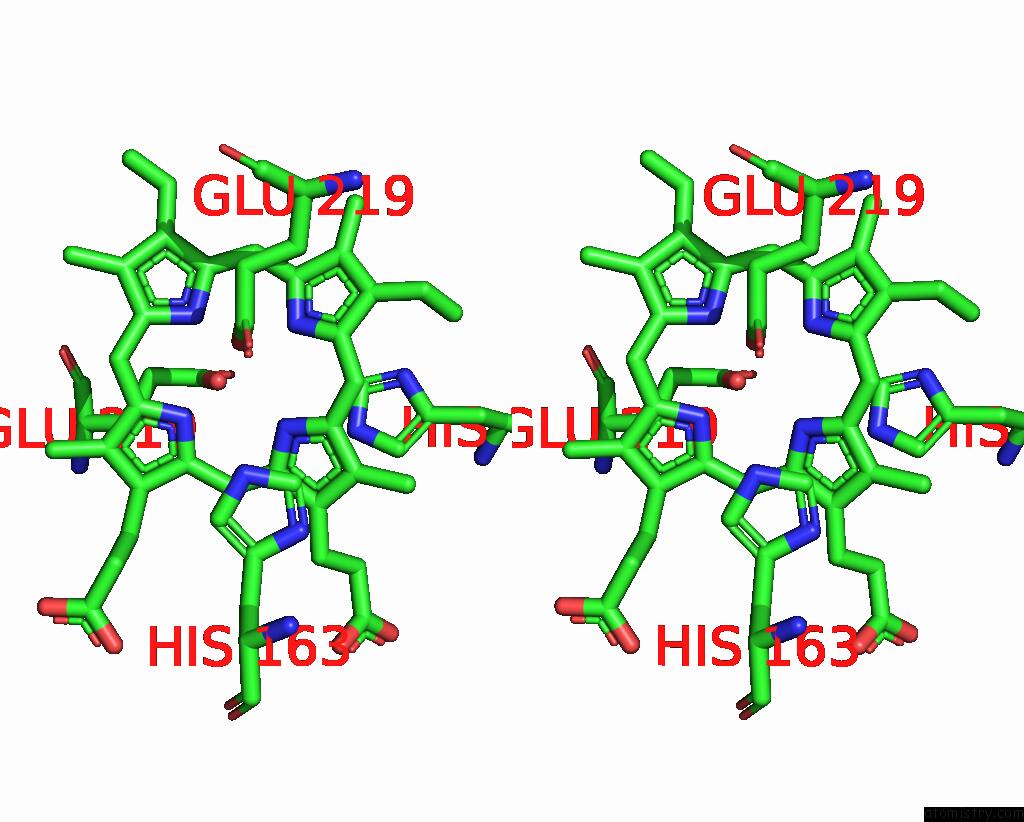
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Heme Exporter in Complex with Mn-Containing Protoporphyrin IX, Mn- Anomalous Data within 5.0Å range:
|
Reference:
H.Nakamura,
T.Hisano,
M.M.Rahman,
T.Tosha,
M.Shirouzu,
Y.Shiro.
Structural Basis For Heme Detoxification By An Atp-Binding Cassette-Type Efflux Pump in Gram-Positive Pathogenic Bacteria. Proc.Natl.Acad.Sci.Usa V. 119 85119 2022.
ISSN: ESSN 1091-6490
PubMed: 35767641
DOI: 10.1073/PNAS.2123385119
Page generated: Sun Oct 6 10:59:15 2024
ISSN: ESSN 1091-6490
PubMed: 35767641
DOI: 10.1073/PNAS.2123385119
Last articles
F in 4H52F in 4H4O
F in 4H4C
F in 4GSY
F in 4H3J
F in 4GXJ
F in 4GVH
F in 4GU9
F in 4GSI
F in 4GPT