Manganese »
PDB 7u80-7utt »
7uch »
Manganese in PDB 7uch: Apra Methyltransferase 1 - Gnat in Complex with MN2+ , Sam, and Di- Methyl-Malonate
Protein crystallography data
The structure of Apra Methyltransferase 1 - Gnat in Complex with MN2+ , Sam, and Di- Methyl-Malonate, PDB code: 7uch
was solved by
M.A.Skiba,
Y.Lao,
J.L.Smith,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.23 / 2.18 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 60.572, 88.283, 135.982, 90, 90, 90 |
| R / Rfree (%) | 19.3 / 24.1 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Apra Methyltransferase 1 - Gnat in Complex with MN2+ , Sam, and Di- Methyl-Malonate
(pdb code 7uch). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total only one binding site of Manganese was determined in the Apra Methyltransferase 1 - Gnat in Complex with MN2+ , Sam, and Di- Methyl-Malonate, PDB code: 7uch:
In total only one binding site of Manganese was determined in the Apra Methyltransferase 1 - Gnat in Complex with MN2+ , Sam, and Di- Methyl-Malonate, PDB code: 7uch:
Manganese binding site 1 out of 1 in 7uch
Go back to
Manganese binding site 1 out
of 1 in the Apra Methyltransferase 1 - Gnat in Complex with MN2+ , Sam, and Di- Methyl-Malonate
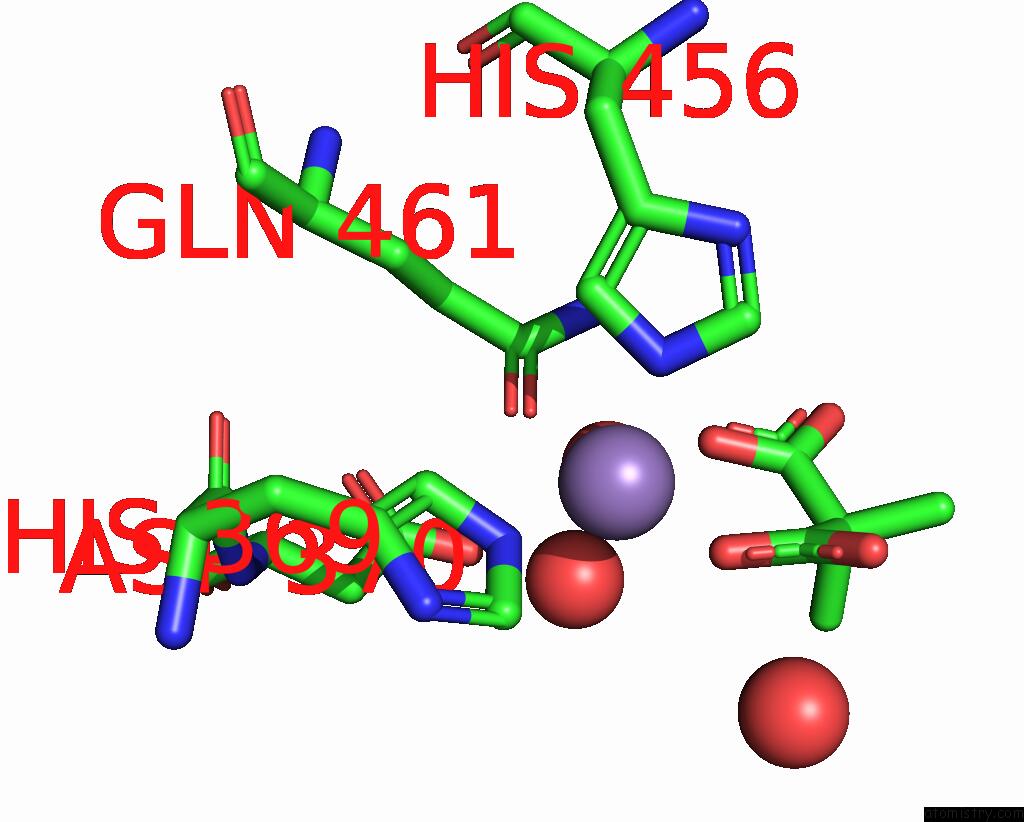
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Apra Methyltransferase 1 - Gnat in Complex with MN2+ , Sam, and Di- Methyl-Malonate within 5.0Å range:
|
Reference:
Y.Lao,
M.A.Skiba,
S.W.Chun,
A.R.H.Narayan,
J.L.Smith.
Structural Basis For Control of Methylation Extent in Polyketide Synthase Metal-Dependent C -Methyltransferases. Acs Chem.Biol. V. 17 2088 2022.
ISSN: ESSN 1554-8937
PubMed: 35594521
DOI: 10.1021/ACSCHEMBIO.2C00085
Page generated: Sun Oct 6 10:48:15 2024
ISSN: ESSN 1554-8937
PubMed: 35594521
DOI: 10.1021/ACSCHEMBIO.2C00085
Last articles
Cl in 7UKTCl in 7UM9
Cl in 7ULA
Cl in 7UL2
Cl in 7UKP
Cl in 7UKO
Cl in 7UKB
Cl in 7UJ2
Cl in 7UJK
Cl in 7UJE