Manganese »
PDB 6rwz-6txf »
6sa1 »
Manganese in PDB 6sa1: Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp
Protein crystallography data
The structure of Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp, PDB code: 6sa1
was solved by
N.C.Brissett,
A.J.Doherty,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 88.66 / 2.01 |
| Space group | P 4 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 198.250, 198.250, 85.300, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.5 / 20.4 |
Other elements in 6sa1:
The structure of Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Manganese Binding Sites:
The binding sites of Manganese atom in the Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp
(pdb code 6sa1). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 4 binding sites of Manganese where determined in the Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp, PDB code: 6sa1:
Jump to Manganese binding site number: 1; 2; 3; 4;
In total 4 binding sites of Manganese where determined in the Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp, PDB code: 6sa1:
Jump to Manganese binding site number: 1; 2; 3; 4;
Manganese binding site 1 out of 4 in 6sa1
Go back to
Manganese binding site 1 out
of 4 in the Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp
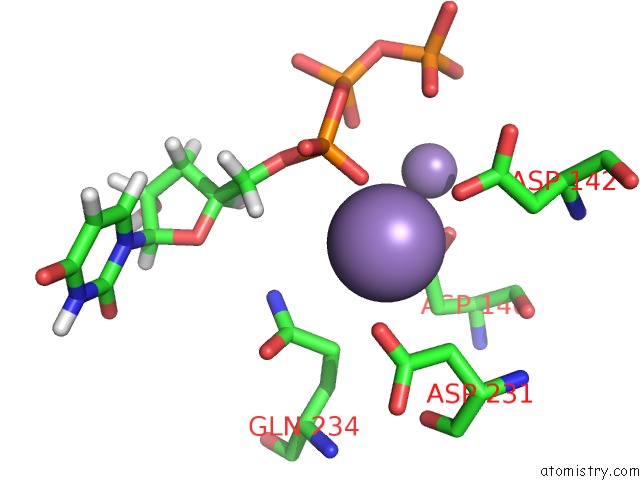
Mono view
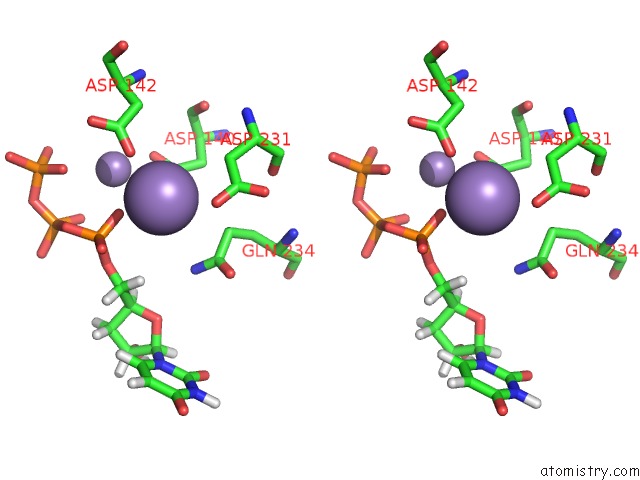
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp within 5.0Å range:
|
Manganese binding site 2 out of 4 in 6sa1
Go back to
Manganese binding site 2 out
of 4 in the Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp
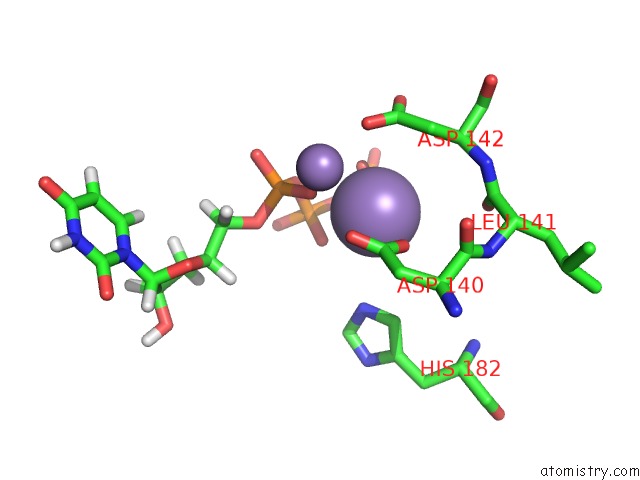
Mono view
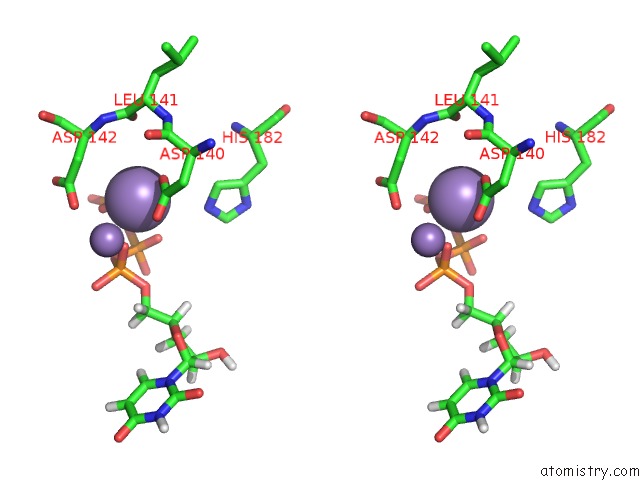
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp within 5.0Å range:
|
Manganese binding site 3 out of 4 in 6sa1
Go back to
Manganese binding site 3 out
of 4 in the Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp
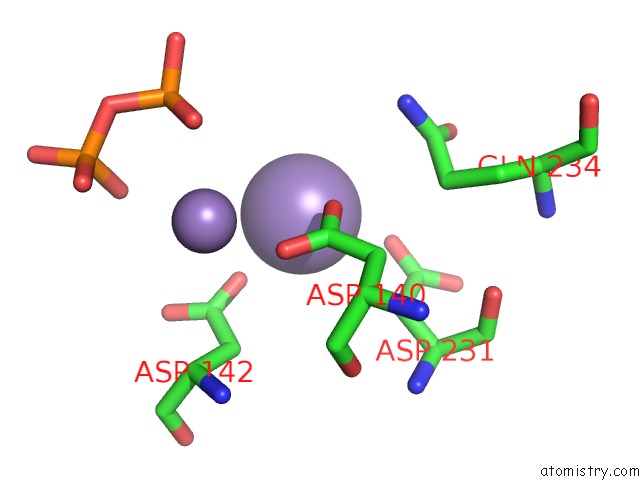
Mono view
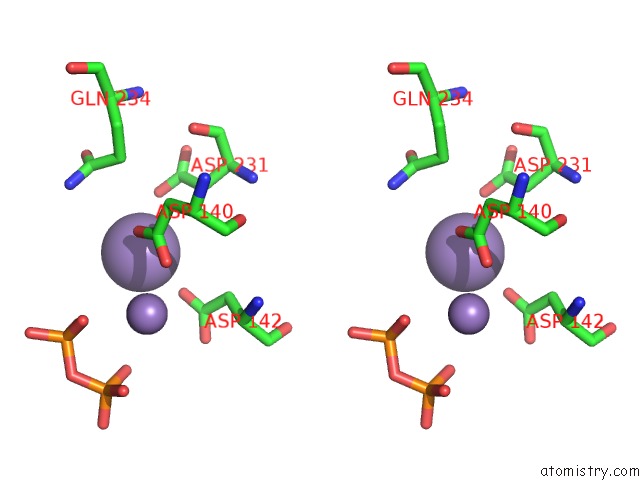
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp within 5.0Å range:
|
Manganese binding site 4 out of 4 in 6sa1
Go back to
Manganese binding site 4 out
of 4 in the Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp
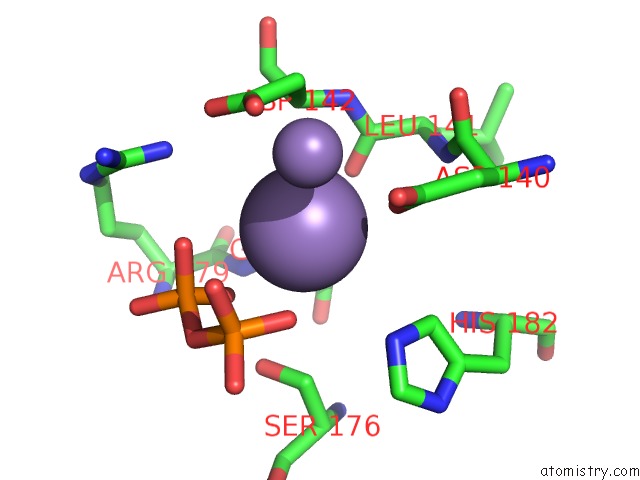
Mono view
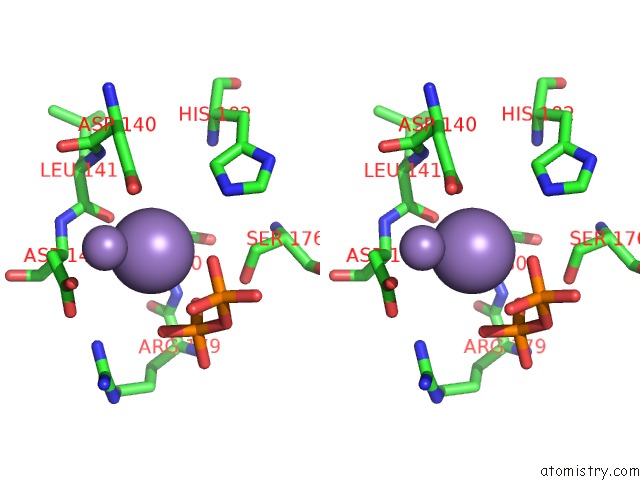
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of Post Catalytic Complex of Prim-Polc From Mycobacterium Smegmatis with Gapped Dna and 3'-Dutp within 5.0Å range:
|
Reference:
N.C.Brissett,
K.Zabrady,
P.Plocinski,
J.Bianchi,
M.Korycka-Machala,
A.Brzostek,
J.Dziadek,
A.J.Doherty.
Molecular Basis For Dna Repair Synthesis on Short Gaps By Mycobacterial Primase-Polymerase C. Nat Commun V. 11 4196 2020.
ISSN: ESSN 2041-1723
PubMed: 32826907
DOI: 10.1038/S41467-020-18012-8
Page generated: Sun Oct 6 07:06:37 2024
ISSN: ESSN 2041-1723
PubMed: 32826907
DOI: 10.1038/S41467-020-18012-8
Last articles
Ca in 5T2HCa in 5SZQ
Ca in 5SZO
Ca in 5SZP
Ca in 5SZN
Ca in 5SZM
Ca in 5SZL
Ca in 5SY1
Ca in 5SWI
Ca in 5SVE