Manganese »
PDB 6qv9-6ru4 »
6qvv »
Manganese in PDB 6qvv: Structure and Function of Phenuiviridae Cap Snatching Endonucleases
Protein crystallography data
The structure of Structure and Function of Phenuiviridae Cap Snatching Endonucleases, PDB code: 6qvv
was solved by
J.Reguera,
R.Jones,
G.Bragagniolo,
S.Lessoued,
M.Mate,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.18 / 2.40 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 101.004, 109.638, 58.159, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.1 / 25.6 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Structure and Function of Phenuiviridae Cap Snatching Endonucleases
(pdb code 6qvv). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 2 binding sites of Manganese where determined in the Structure and Function of Phenuiviridae Cap Snatching Endonucleases, PDB code: 6qvv:
Jump to Manganese binding site number: 1; 2;
In total 2 binding sites of Manganese where determined in the Structure and Function of Phenuiviridae Cap Snatching Endonucleases, PDB code: 6qvv:
Jump to Manganese binding site number: 1; 2;
Manganese binding site 1 out of 2 in 6qvv
Go back to
Manganese binding site 1 out
of 2 in the Structure and Function of Phenuiviridae Cap Snatching Endonucleases
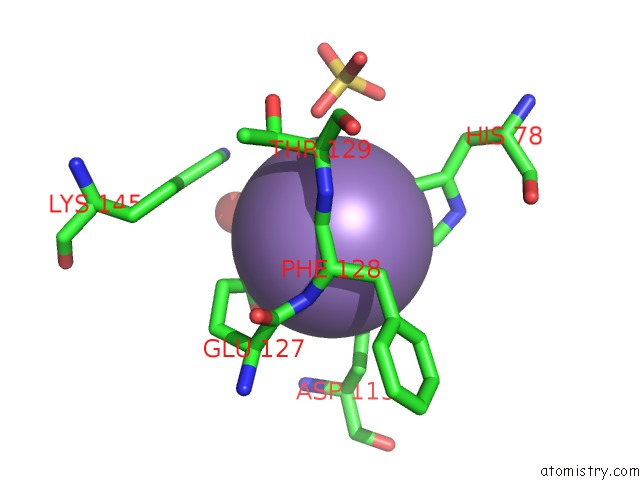
Mono view
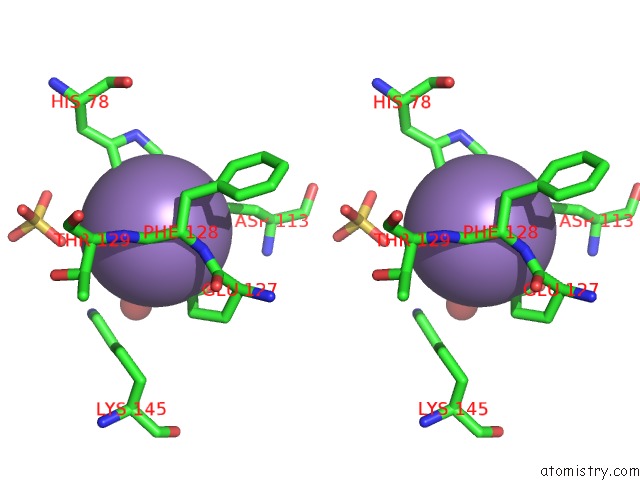
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Structure and Function of Phenuiviridae Cap Snatching Endonucleases within 5.0Å range:
|
Manganese binding site 2 out of 2 in 6qvv
Go back to
Manganese binding site 2 out
of 2 in the Structure and Function of Phenuiviridae Cap Snatching Endonucleases
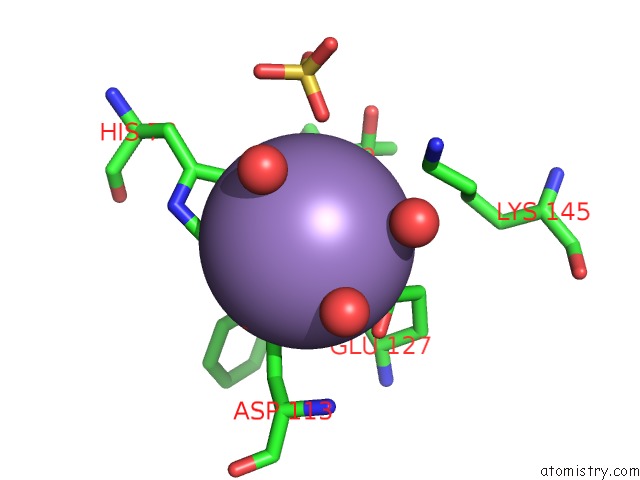
Mono view
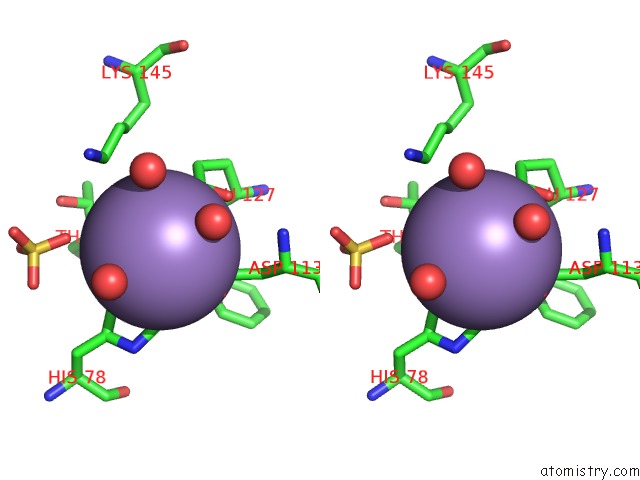
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Structure and Function of Phenuiviridae Cap Snatching Endonucleases within 5.0Å range:
|
Reference:
R.Jones,
S.Lessoued,
K.Meier,
S.Devignot,
S.Barata-Garcia,
M.Mate,
G.Bragagnolo,
F.Weber,
M.Rosenthal,
J.Reguera.
Structure and Function of the Toscana Virus Cap-Snatching Endonuclease. Nucleic Acids Res. V. 47 10914 2019.
ISSN: ESSN 1362-4962
PubMed: 31584100
DOI: 10.1093/NAR/GKZ838
Page generated: Sun Oct 6 06:56:21 2024
ISSN: ESSN 1362-4962
PubMed: 31584100
DOI: 10.1093/NAR/GKZ838
Last articles
Ca in 5W0RCa in 5VYG
Ca in 5VT8
Ca in 5VYF
Ca in 5VYB
Ca in 5VXZ
Ca in 5VTM
Ca in 5VWM
Ca in 5VTD
Ca in 5VUG