Manganese »
PDB 6oe2-6q9f »
6pxu »
Manganese in PDB 6pxu: Crystal Structure of Human Galnac-T12 Bound to A Diglycosylated Peptide, MN2+, and Udp
Enzymatic activity of Crystal Structure of Human Galnac-T12 Bound to A Diglycosylated Peptide, MN2+, and Udp
All present enzymatic activity of Crystal Structure of Human Galnac-T12 Bound to A Diglycosylated Peptide, MN2+, and Udp:
2.4.1.41;
2.4.1.41;
Protein crystallography data
The structure of Crystal Structure of Human Galnac-T12 Bound to A Diglycosylated Peptide, MN2+, and Udp, PDB code: 6pxu
was solved by
N.L.Samara,
A.J.Fernandez,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.03 / 2.01 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 72.820, 73.123, 74.441, 113.08, 100.50, 108.23 |
| R / Rfree (%) | 17.1 / 21.9 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Crystal Structure of Human Galnac-T12 Bound to A Diglycosylated Peptide, MN2+, and Udp
(pdb code 6pxu). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 4 binding sites of Manganese where determined in the Crystal Structure of Human Galnac-T12 Bound to A Diglycosylated Peptide, MN2+, and Udp, PDB code: 6pxu:
Jump to Manganese binding site number: 1; 2; 3; 4;
In total 4 binding sites of Manganese where determined in the Crystal Structure of Human Galnac-T12 Bound to A Diglycosylated Peptide, MN2+, and Udp, PDB code: 6pxu:
Jump to Manganese binding site number: 1; 2; 3; 4;
Manganese binding site 1 out of 4 in 6pxu
Go back to
Manganese binding site 1 out
of 4 in the Crystal Structure of Human Galnac-T12 Bound to A Diglycosylated Peptide, MN2+, and Udp
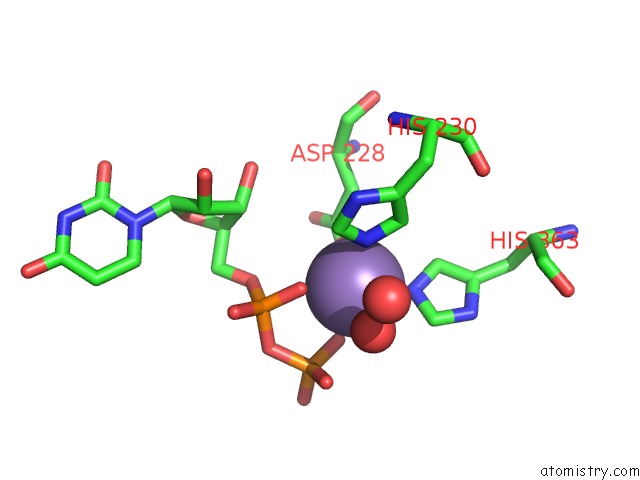
Mono view
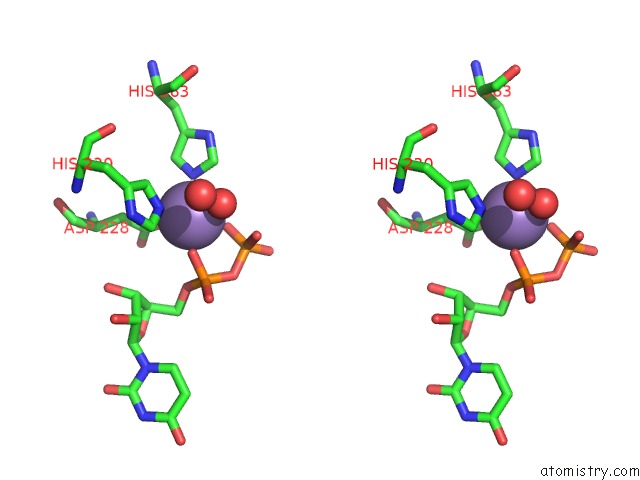
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Crystal Structure of Human Galnac-T12 Bound to A Diglycosylated Peptide, MN2+, and Udp within 5.0Å range:
|
Manganese binding site 2 out of 4 in 6pxu
Go back to
Manganese binding site 2 out
of 4 in the Crystal Structure of Human Galnac-T12 Bound to A Diglycosylated Peptide, MN2+, and Udp
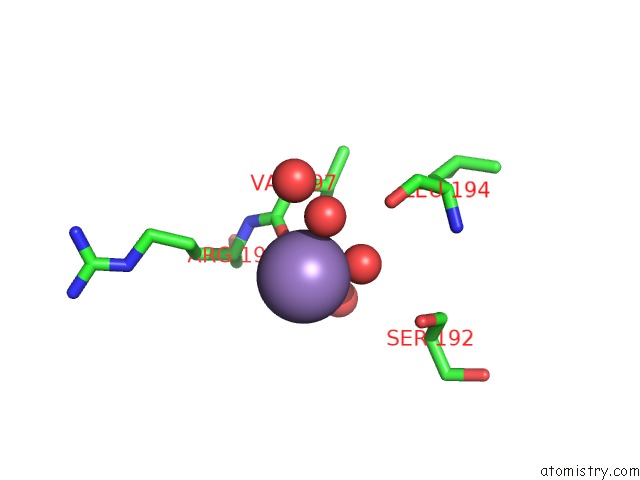
Mono view
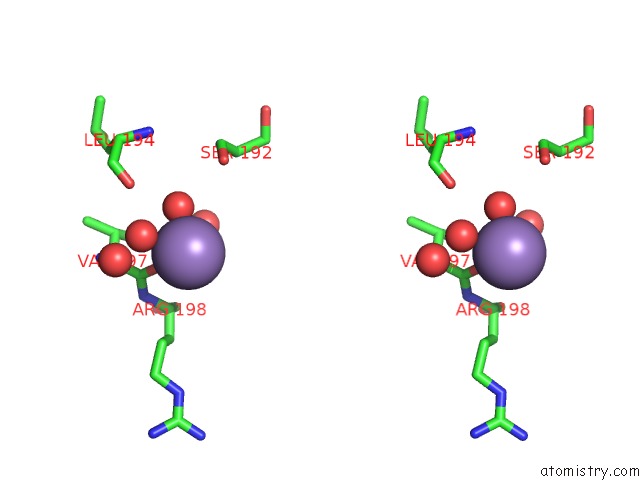
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Crystal Structure of Human Galnac-T12 Bound to A Diglycosylated Peptide, MN2+, and Udp within 5.0Å range:
|
Manganese binding site 3 out of 4 in 6pxu
Go back to
Manganese binding site 3 out
of 4 in the Crystal Structure of Human Galnac-T12 Bound to A Diglycosylated Peptide, MN2+, and Udp
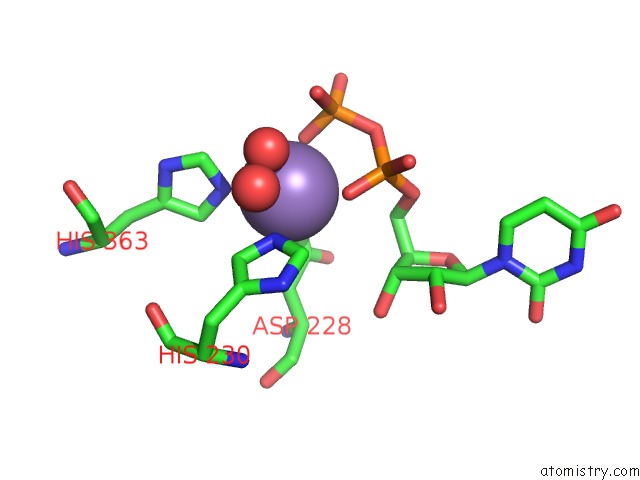
Mono view
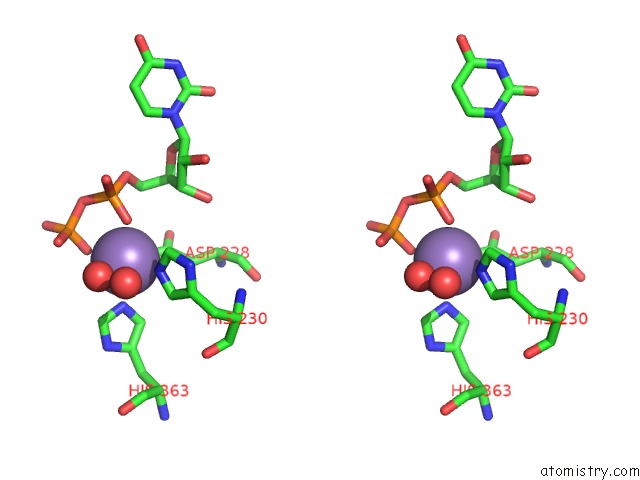
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Crystal Structure of Human Galnac-T12 Bound to A Diglycosylated Peptide, MN2+, and Udp within 5.0Å range:
|
Manganese binding site 4 out of 4 in 6pxu
Go back to
Manganese binding site 4 out
of 4 in the Crystal Structure of Human Galnac-T12 Bound to A Diglycosylated Peptide, MN2+, and Udp
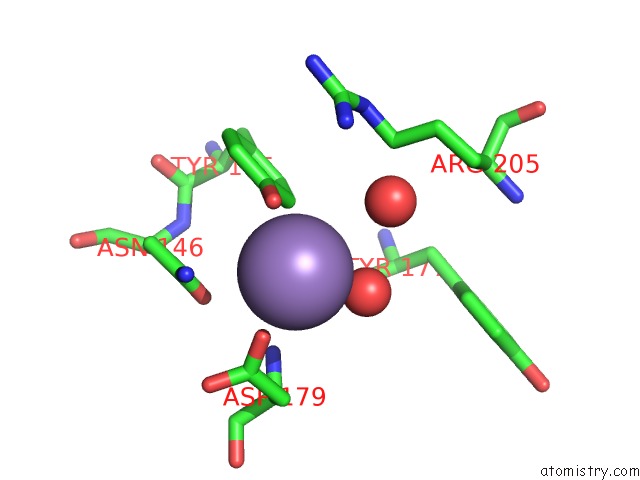
Mono view
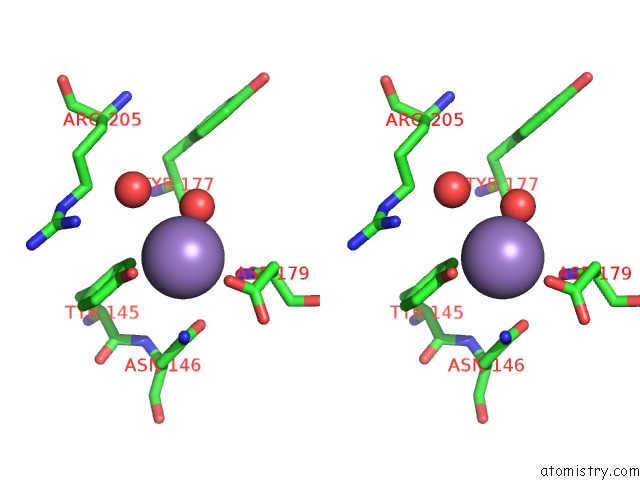
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of Crystal Structure of Human Galnac-T12 Bound to A Diglycosylated Peptide, MN2+, and Udp within 5.0Å range:
|
Reference:
A.J.Fernandez,
E.J.P.Daniel,
S.P.Mahajan,
J.J.Gray,
T.A.Gerken,
L.A.Tabak,
N.L.Samara.
The Structure of the Colorectal Cancer-Associated Enzyme Galnac-T12 Reveals How Nonconserved Residues Dictate Its Function. Proc.Natl.Acad.Sci.Usa V. 116 20404 2019.
ISSN: ESSN 1091-6490
PubMed: 31548401
DOI: 10.1073/PNAS.1902211116
Page generated: Sun Oct 6 05:52:42 2024
ISSN: ESSN 1091-6490
PubMed: 31548401
DOI: 10.1073/PNAS.1902211116
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO