Manganese »
PDB 6oe2-6q9f »
6ov6 »
Manganese in PDB 6ov6: Crystallographic Structure of the C24 Protein From the Antarctic Microorganism Bizionia Argentinensis
Protein crystallography data
The structure of Crystallographic Structure of the C24 Protein From the Antarctic Microorganism Bizionia Argentinensis, PDB code: 6ov6
was solved by
S.Klinke,
J.Rinaldi,
B.G.Guimaraes,
L.Pellizza,
M.Aran,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 44.12 / 1.82 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 63.130, 65.250, 239.530, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.8 / 23 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Crystallographic Structure of the C24 Protein From the Antarctic Microorganism Bizionia Argentinensis
(pdb code 6ov6). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total only one binding site of Manganese was determined in the Crystallographic Structure of the C24 Protein From the Antarctic Microorganism Bizionia Argentinensis, PDB code: 6ov6:
In total only one binding site of Manganese was determined in the Crystallographic Structure of the C24 Protein From the Antarctic Microorganism Bizionia Argentinensis, PDB code: 6ov6:
Manganese binding site 1 out of 1 in 6ov6
Go back to
Manganese binding site 1 out
of 1 in the Crystallographic Structure of the C24 Protein From the Antarctic Microorganism Bizionia Argentinensis
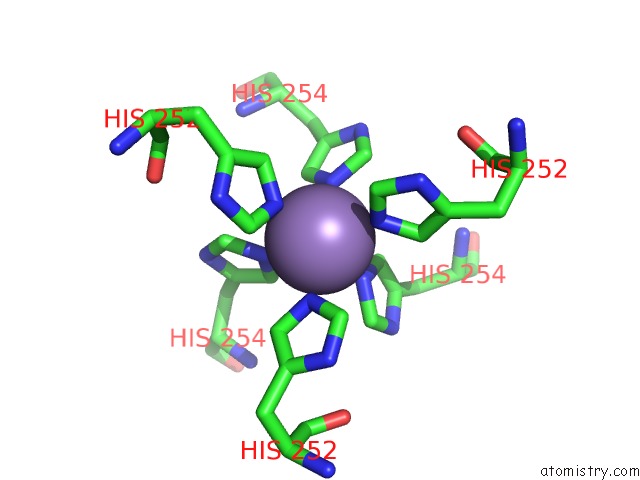
Mono view
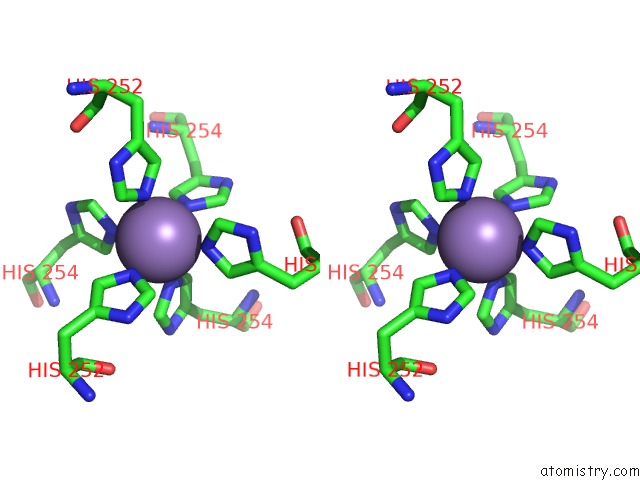
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Crystallographic Structure of the C24 Protein From the Antarctic Microorganism Bizionia Argentinensis within 5.0Å range:
|
Reference:
L.Pellizza,
J.L.Lopez,
S.Vazquez,
G.Sycz,
B.G.Guimaraes,
J.Rinaldi,
F.A.Goldbaum,
M.Aran,
W.P.Mac Cormack,
S.Klinke.
Structure of the Putative Long Tail Fiber Receptor-Binding Tip of A Novel Temperate Bacteriophage From the Antarctic Bacterium Bizionia Argentinensis JUB59. J.Struct.Biol. V. 212 07595 2020.
ISSN: ESSN 1095-8657
PubMed: 32736071
DOI: 10.1016/J.JSB.2020.107595
Page generated: Sun Oct 6 05:46:55 2024
ISSN: ESSN 1095-8657
PubMed: 32736071
DOI: 10.1016/J.JSB.2020.107595
Last articles
Cl in 3DEPCl in 3DEJ
Cl in 3DCL
Cl in 3DDW
Cl in 3DEE
Cl in 3DDH
Cl in 3DDU
Cl in 3DCX
Cl in 3DCU
Cl in 3D53