Manganese »
PDB 6e4q-6f4p »
6ezm »
Manganese in PDB 6ezm: Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae
Enzymatic activity of Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae
All present enzymatic activity of Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae:
4.2.1.19;
4.2.1.19;
Manganese Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 20; Page 3, Binding sites: 21 - 30; Page 4, Binding sites: 31 - 40; Page 5, Binding sites: 41 - 48;Binding sites:
The binding sites of Manganese atom in the Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae (pdb code 6ezm). This binding sites where shown within 5.0 Angstroms radius around Manganese atom.In total 48 binding sites of Manganese where determined in the Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae, PDB code: 6ezm:
Jump to Manganese binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Manganese binding site 1 out of 48 in 6ezm
Go back to
Manganese binding site 1 out
of 48 in the Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae
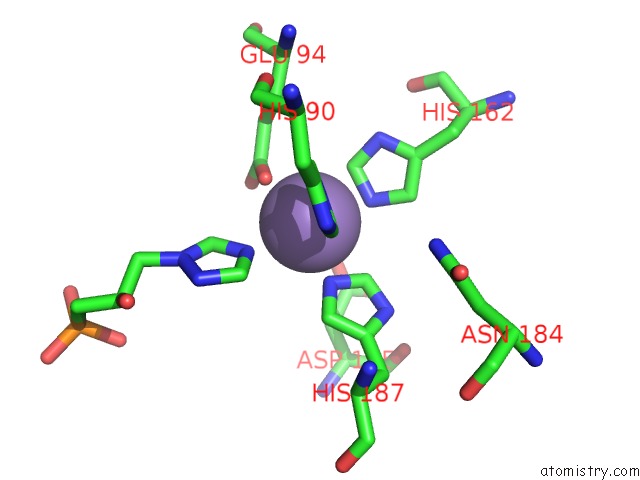
Mono view
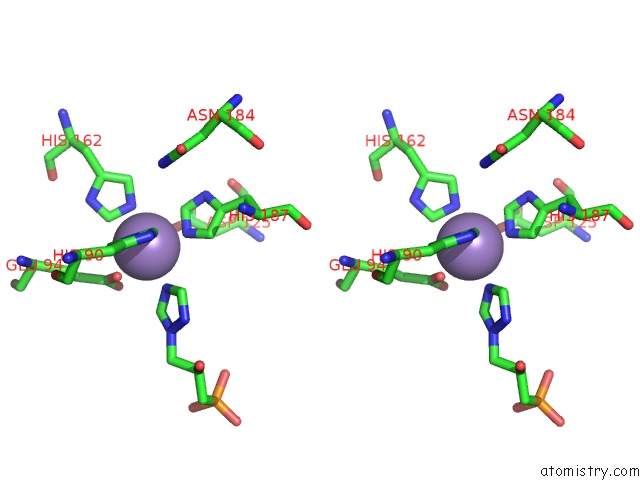
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae within 5.0Å range:
|
Manganese binding site 2 out of 48 in 6ezm
Go back to
Manganese binding site 2 out
of 48 in the Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae
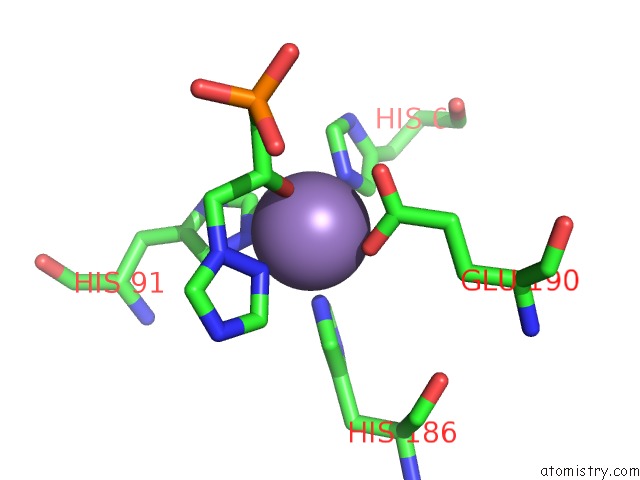
Mono view
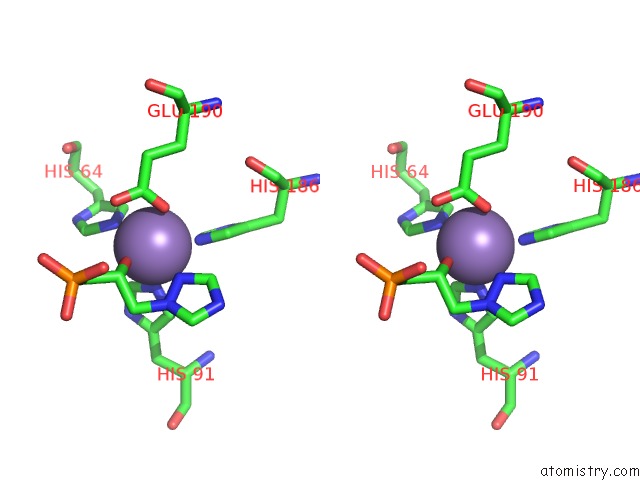
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae within 5.0Å range:
|
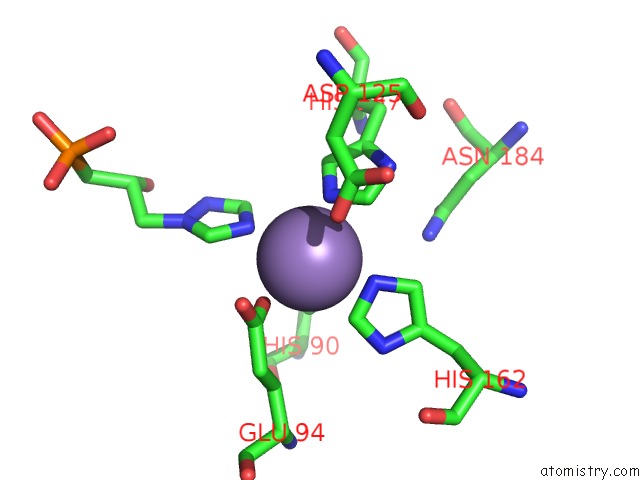
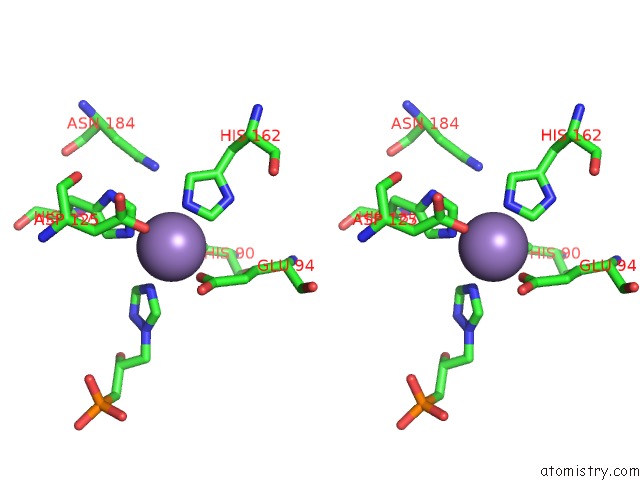
Manganese binding site 3 out of 48 in 6ezm
Go back to
Manganese binding site 3 out
of 48 in the Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae within 5.0Å range:
|
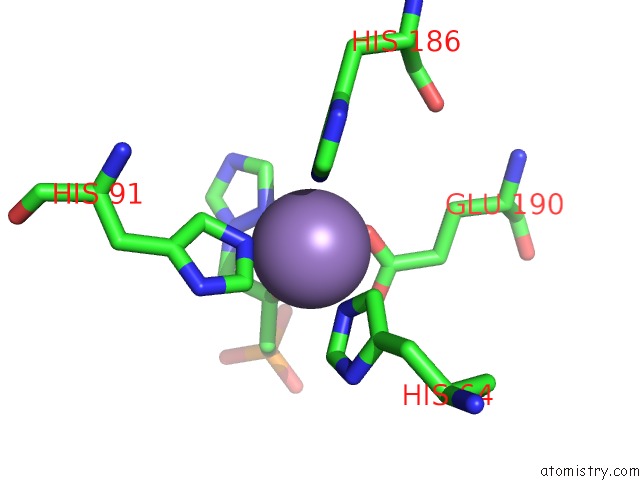
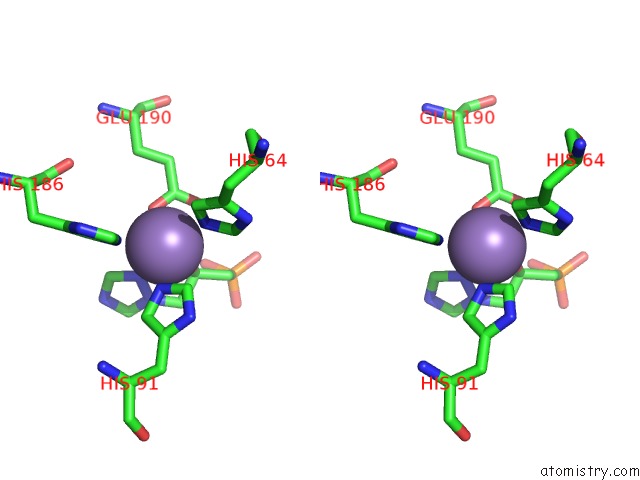
Manganese binding site 4 out of 48 in 6ezm
Go back to
Manganese binding site 4 out
of 48 in the Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae within 5.0Å range:
|
Manganese binding site 5 out of 48 in 6ezm
Go back to
Manganese binding site 5 out
of 48 in the Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae
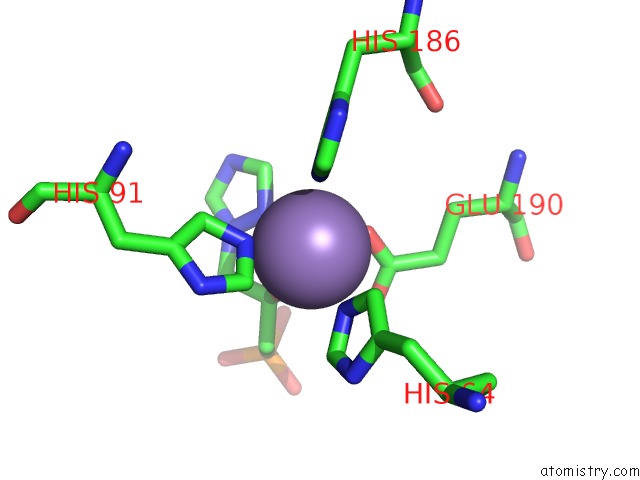
Mono view
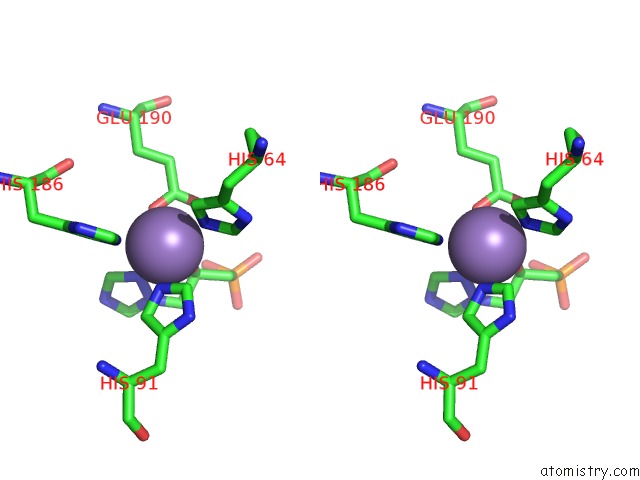
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 5 of Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae within 5.0Å range:
|
Manganese binding site 6 out of 48 in 6ezm
Go back to
Manganese binding site 6 out
of 48 in the Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae
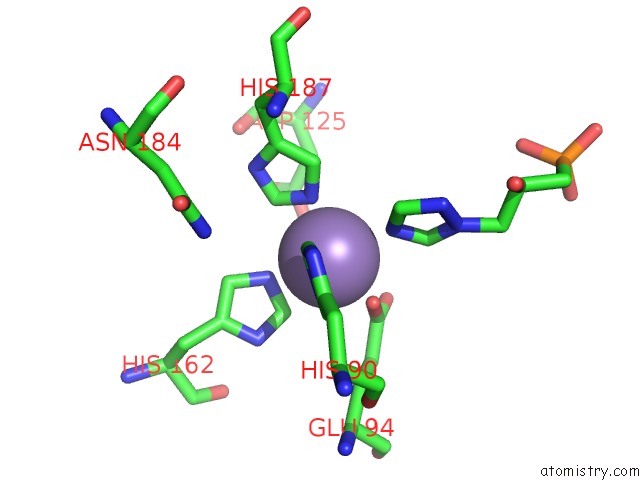
Mono view
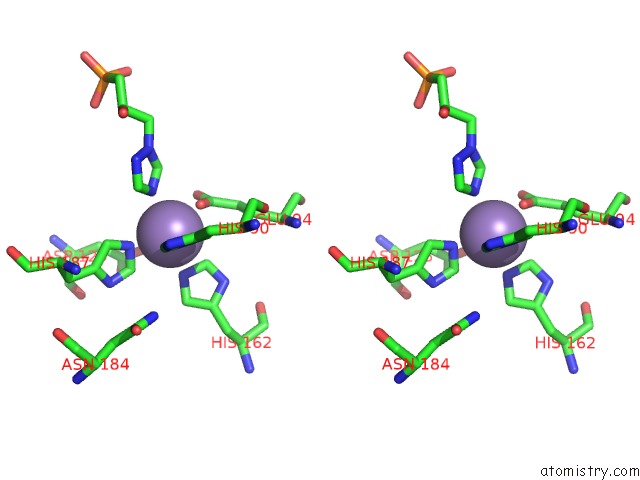
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 6 of Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae within 5.0Å range:
|
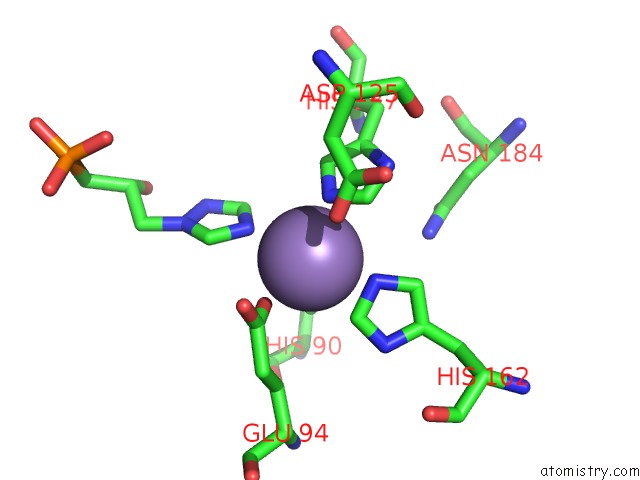
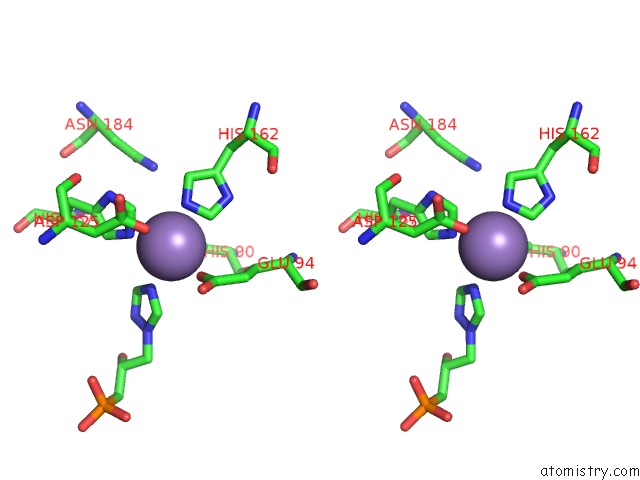
Manganese binding site 7 out of 48 in 6ezm
Go back to
Manganese binding site 7 out
of 48 in the Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 7 of Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae within 5.0Å range:
|
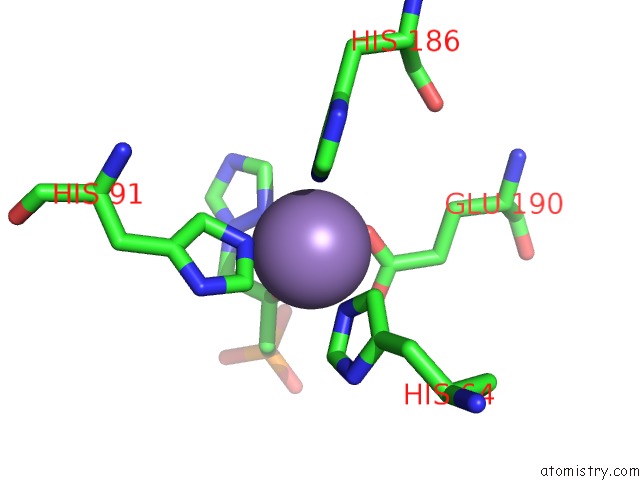
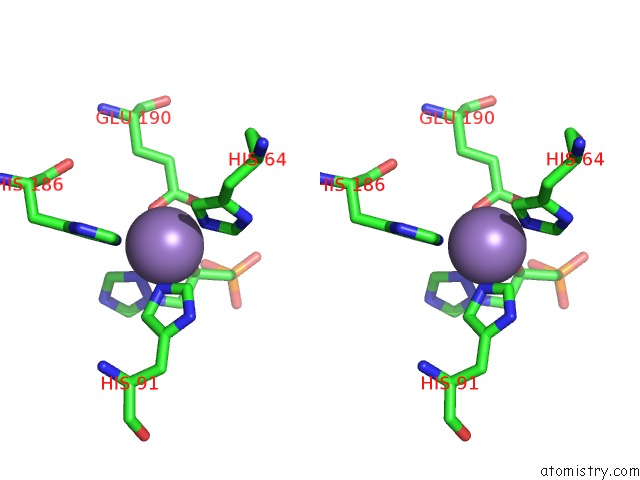
Manganese binding site 8 out of 48 in 6ezm
Go back to
Manganese binding site 8 out
of 48 in the Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 8 of Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae within 5.0Å range:
|
Manganese binding site 9 out of 48 in 6ezm
Go back to
Manganese binding site 9 out
of 48 in the Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae
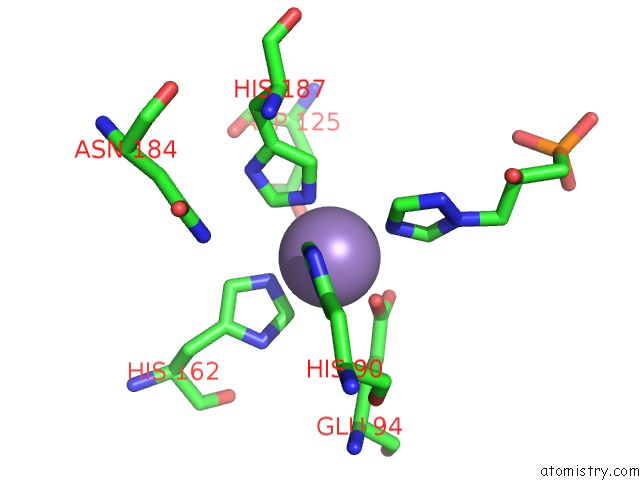
Mono view
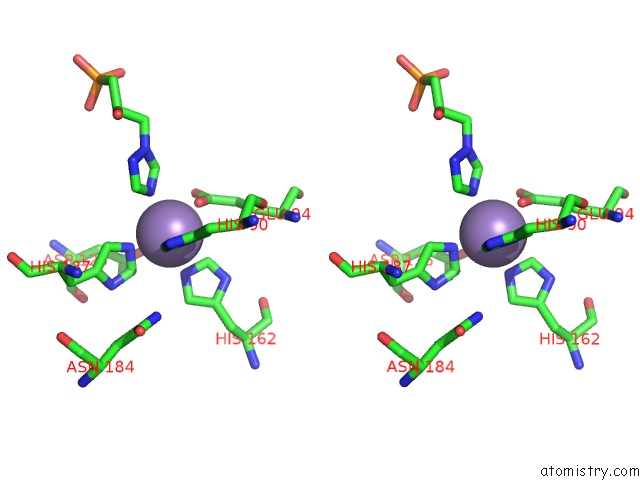
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 9 of Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae within 5.0Å range:
|
Manganese binding site 10 out of 48 in 6ezm
Go back to
Manganese binding site 10 out
of 48 in the Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae
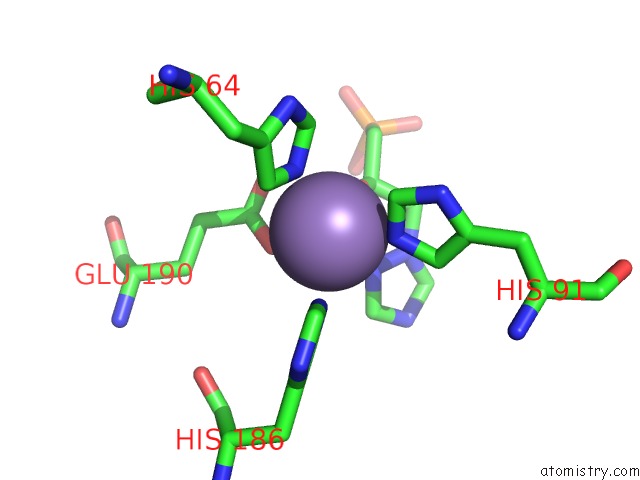
Mono view
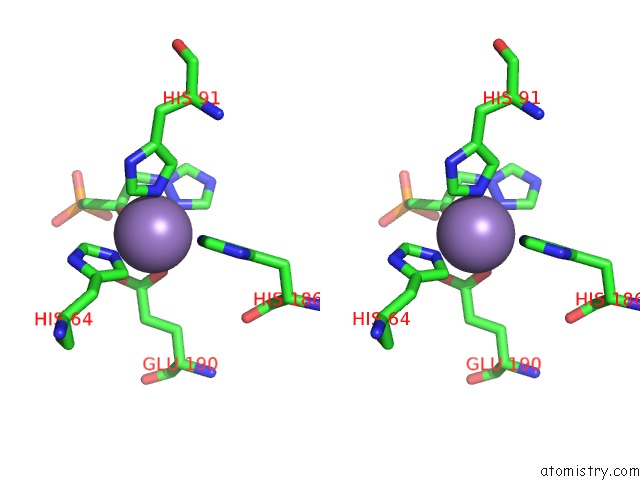
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 10 of Imidazoleglycerol-Phosphate Dehydratase From Saccharomyces Cerevisiae within 5.0Å range:
|
Reference:
S.Rawson,
C.Bisson,
D.L.Hurdiss,
A.Fazal,
M.J.Mcphillie,
S.E.Sedelnikova,
P.J.Baker,
D.W.Rice,
S.P.Muench.
Elucidating the Structural Basis For Differing Enzyme Inhibitor Potency By Cryo-Em. Proc. Natl. Acad. Sci. V. 115 1795 2018U.S.A..
ISSN: ESSN 1091-6490
PubMed: 29434040
DOI: 10.1073/PNAS.1708839115
Page generated: Sun Oct 6 04:22:10 2024
ISSN: ESSN 1091-6490
PubMed: 29434040
DOI: 10.1073/PNAS.1708839115
Last articles
F in 7MIGF in 7MLD
F in 7MKX
F in 7MGK
F in 7MGJ
F in 7MHD
F in 7MFH
F in 7MHC
F in 7MGE
F in 7MFD