Manganese »
PDB 5wly-5yvs »
5yl3 »
Manganese in PDB 5yl3: Crystal Structure of Horse Heart Myoglobin Reconstituted with Manganese Porphycene in Resting State at pH 8.5
Protein crystallography data
The structure of Crystal Structure of Horse Heart Myoglobin Reconstituted with Manganese Porphycene in Resting State at pH 8.5, PDB code: 5yl3
was solved by
K.Oohora,
H.Meichin,
Y.Kihira,
H.Sugimoto,
Y.Shiro,
T.Hayashi,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 1.50 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 34.575, 28.706, 62.702, 90.00, 106.15, 90.00 |
| R / Rfree (%) | 16.8 / 20.8 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Crystal Structure of Horse Heart Myoglobin Reconstituted with Manganese Porphycene in Resting State at pH 8.5
(pdb code 5yl3). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total only one binding site of Manganese was determined in the Crystal Structure of Horse Heart Myoglobin Reconstituted with Manganese Porphycene in Resting State at pH 8.5, PDB code: 5yl3:
In total only one binding site of Manganese was determined in the Crystal Structure of Horse Heart Myoglobin Reconstituted with Manganese Porphycene in Resting State at pH 8.5, PDB code: 5yl3:
Manganese binding site 1 out of 1 in 5yl3
Go back to
Manganese binding site 1 out
of 1 in the Crystal Structure of Horse Heart Myoglobin Reconstituted with Manganese Porphycene in Resting State at pH 8.5
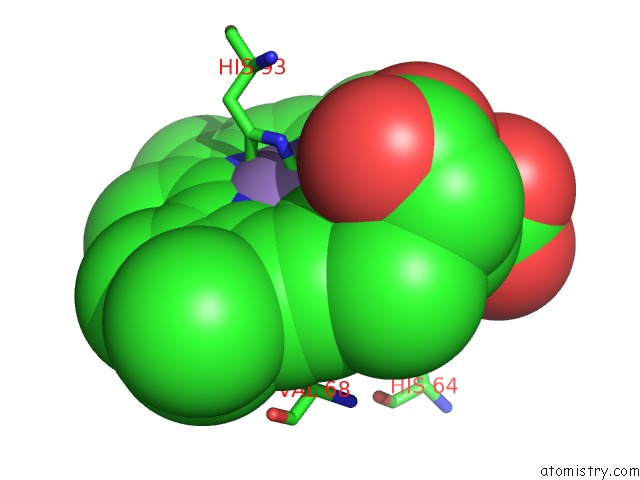
Mono view
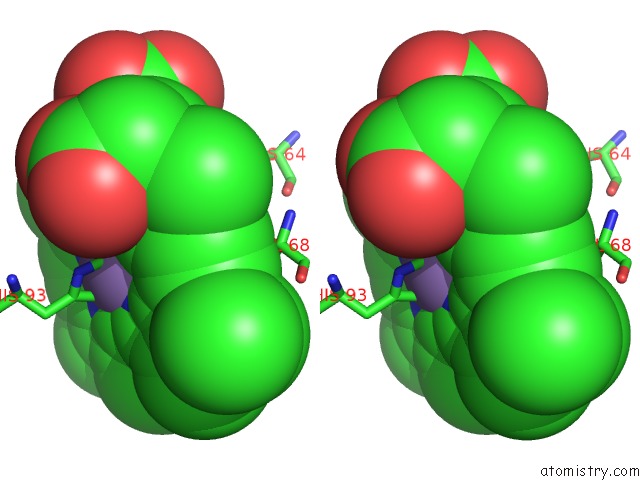
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Crystal Structure of Horse Heart Myoglobin Reconstituted with Manganese Porphycene in Resting State at pH 8.5 within 5.0Å range:
|
Reference:
K.Oohora,
H.Meichin,
Y.Kihira,
H.Sugimoto,
Y.Shiro,
T.Hayashi.
Manganese(V) Porphycene Complex Responsible For Inert C-H Bond Hydroxylation in A Myoglobin Matrix. J. Am. Chem. Soc. V. 139 18460 2017.
ISSN: ESSN 1520-5126
PubMed: 29237270
DOI: 10.1021/JACS.7B11288
Page generated: Sat Aug 16 19:38:56 2025
ISSN: ESSN 1520-5126
PubMed: 29237270
DOI: 10.1021/JACS.7B11288
Last articles
Na in 1D7VNa in 1D7U
Na in 1D7S
Na in 1D6W
Na in 1D3Y
Na in 1D4P
Na in 1D7R
Na in 1D3T
Na in 1D3Q
Na in 1D3D