Manganese »
PDB 4qsh-4u3o »
4r89 »
Manganese in PDB 4r89: Crystal Structure of PAFAN1 - 5' Flap Dna Complex with Manganase
Protein crystallography data
The structure of Crystal Structure of PAFAN1 - 5' Flap Dna Complex with Manganase, PDB code: 4r89
was solved by
Y.Cho,
G.H.Gwon,
Y.R.Kim,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 44.64 / 4.00 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 81.115, 106.944, 107.541, 90.00, 89.84, 90.00 |
| R / Rfree (%) | 22.4 / 27.8 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Crystal Structure of PAFAN1 - 5' Flap Dna Complex with Manganase
(pdb code 4r89). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 4 binding sites of Manganese where determined in the Crystal Structure of PAFAN1 - 5' Flap Dna Complex with Manganase, PDB code: 4r89:
Jump to Manganese binding site number: 1; 2; 3; 4;
In total 4 binding sites of Manganese where determined in the Crystal Structure of PAFAN1 - 5' Flap Dna Complex with Manganase, PDB code: 4r89:
Jump to Manganese binding site number: 1; 2; 3; 4;
Manganese binding site 1 out of 4 in 4r89
Go back to
Manganese binding site 1 out
of 4 in the Crystal Structure of PAFAN1 - 5' Flap Dna Complex with Manganase
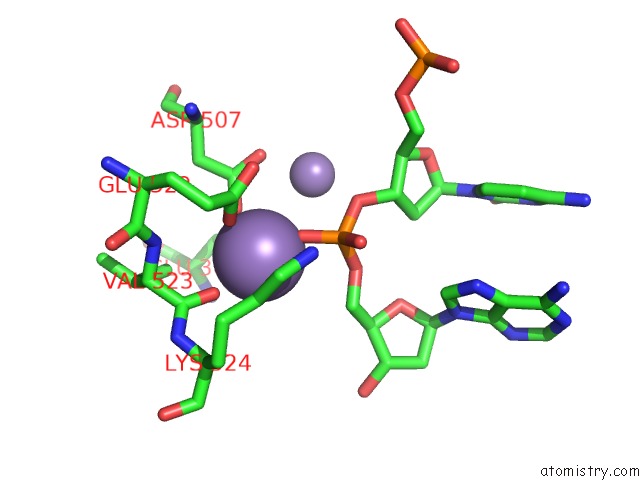
Mono view
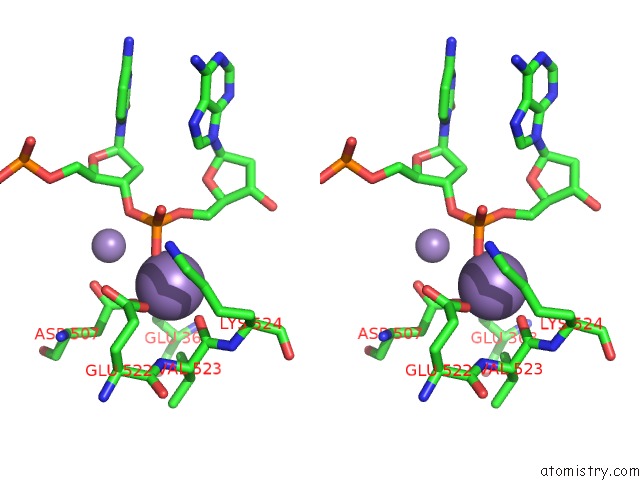
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Crystal Structure of PAFAN1 - 5' Flap Dna Complex with Manganase within 5.0Å range:
|
Manganese binding site 2 out of 4 in 4r89
Go back to
Manganese binding site 2 out
of 4 in the Crystal Structure of PAFAN1 - 5' Flap Dna Complex with Manganase
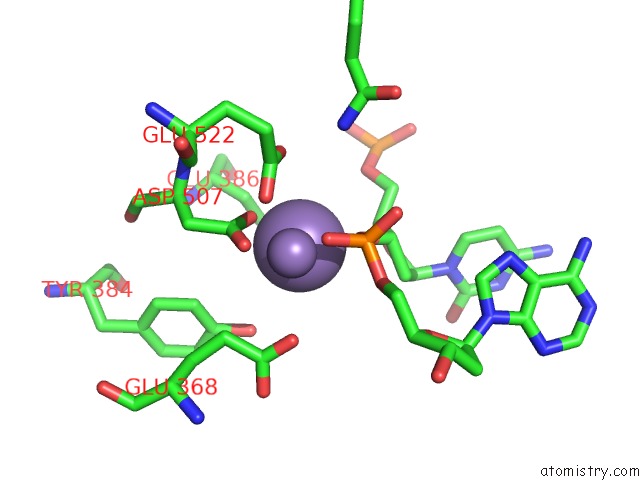
Mono view
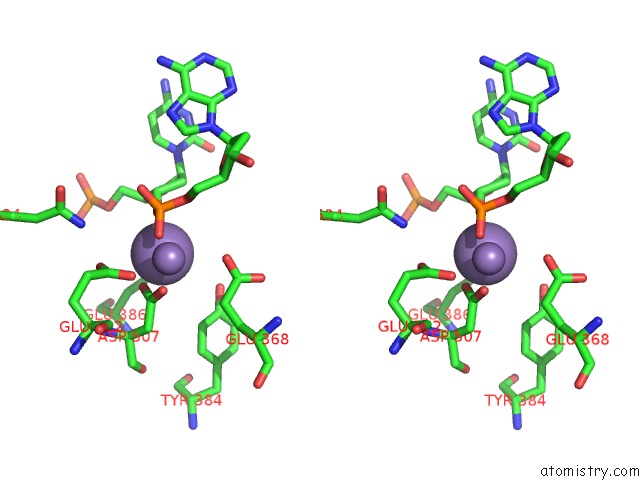
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Crystal Structure of PAFAN1 - 5' Flap Dna Complex with Manganase within 5.0Å range:
|
Manganese binding site 3 out of 4 in 4r89
Go back to
Manganese binding site 3 out
of 4 in the Crystal Structure of PAFAN1 - 5' Flap Dna Complex with Manganase
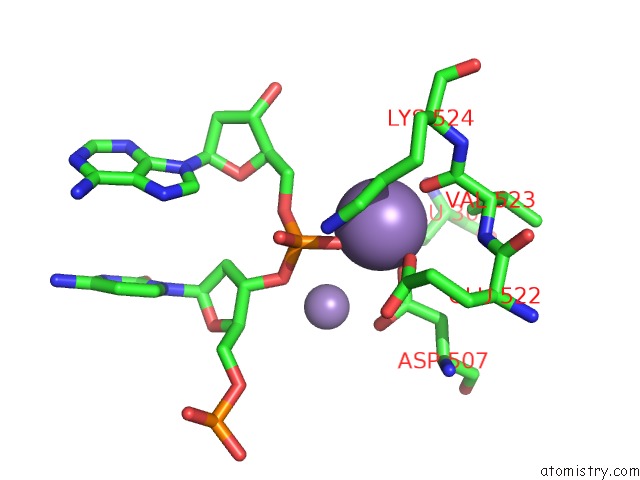
Mono view
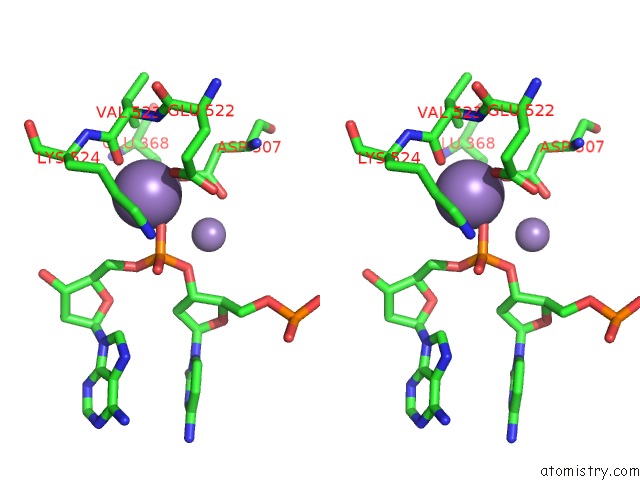
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Crystal Structure of PAFAN1 - 5' Flap Dna Complex with Manganase within 5.0Å range:
|
Manganese binding site 4 out of 4 in 4r89
Go back to
Manganese binding site 4 out
of 4 in the Crystal Structure of PAFAN1 - 5' Flap Dna Complex with Manganase
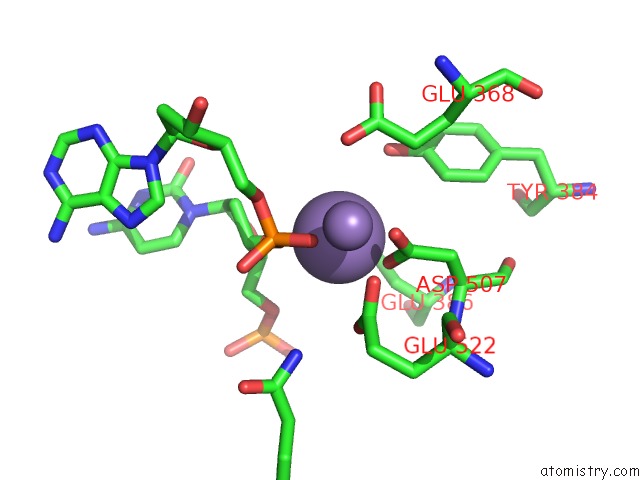
Mono view
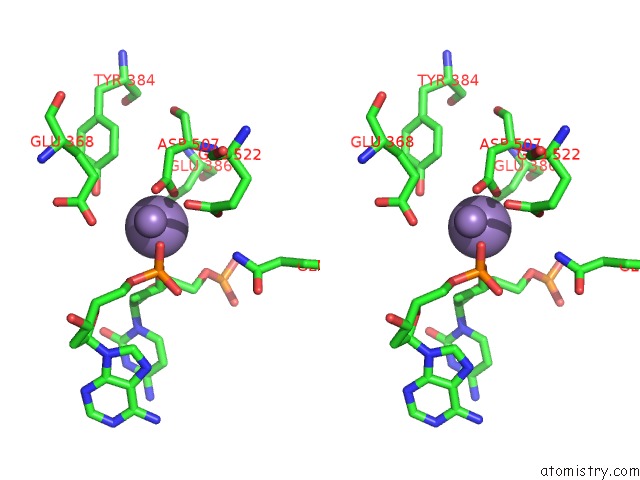
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of Crystal Structure of PAFAN1 - 5' Flap Dna Complex with Manganase within 5.0Å range:
|
Reference:
G.H.Gwon,
Y.R.Kim,
Y.Liu,
A.T.Watson,
A.Jo,
T.J.Etheridge,
F.Yuan,
Y.Zhang,
Y.C.Kim,
A.M.Carr,
Y.Cho.
Crystal Structure of A Fanconi Anemia-Associated Nuclease Homolog Bound to 5' Flap Dna: Basis of Interstrand Cross-Link Repair By FAN1 Genes Dev. V. 28 2276 2014.
ISSN: ISSN 0890-9369
PubMed: 25319828
DOI: 10.1101/GAD.248492.114
Page generated: Sat Oct 5 21:07:07 2024
ISSN: ISSN 0890-9369
PubMed: 25319828
DOI: 10.1101/GAD.248492.114
Last articles
Cl in 5WNZCl in 5WNX
Cl in 5WNM
Cl in 5WNK
Cl in 5WNL
Cl in 5WNJ
Cl in 5WNI
Cl in 5WMT
Cl in 5WMP
Cl in 5WN3