Manganese »
PDB 4mu4-4nx7 »
4nfw »
Manganese in PDB 4nfw: Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli
Protein crystallography data
The structure of Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli, PDB code: 4nfw
was solved by
M.K.Hong,
J.K.Kim,
L.W.Kang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 32.42 / 2.30 |
| Space group | P 41 |
| Cell size a, b, c (Å), α, β, γ (°) | 111.387, 111.387, 247.489, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 24 / 28.9 |
Manganese Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 20; Page 3, Binding sites: 21 - 22;Binding sites:
The binding sites of Manganese atom in the Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli (pdb code 4nfw). This binding sites where shown within 5.0 Angstroms radius around Manganese atom.In total 22 binding sites of Manganese where determined in the Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli, PDB code: 4nfw:
Jump to Manganese binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Manganese binding site 1 out of 22 in 4nfw
Go back to
Manganese binding site 1 out
of 22 in the Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli
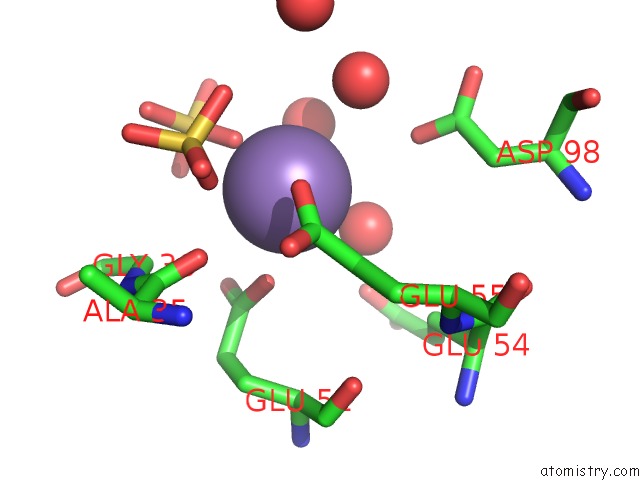
Mono view
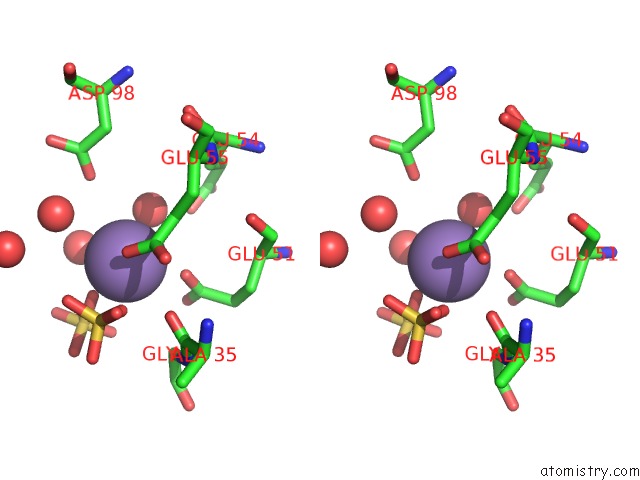
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli within 5.0Å range:
|
Manganese binding site 2 out of 22 in 4nfw
Go back to
Manganese binding site 2 out
of 22 in the Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli
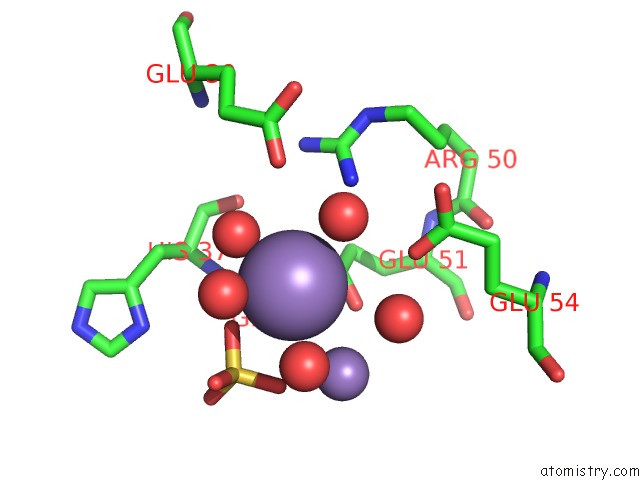
Mono view
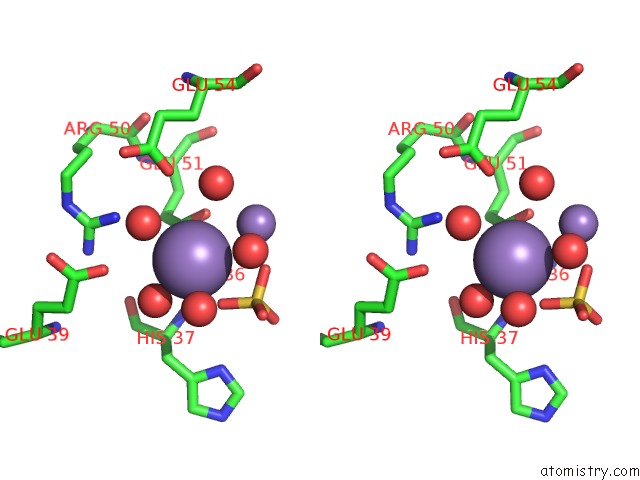
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli within 5.0Å range:
|
Manganese binding site 3 out of 22 in 4nfw
Go back to
Manganese binding site 3 out
of 22 in the Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli
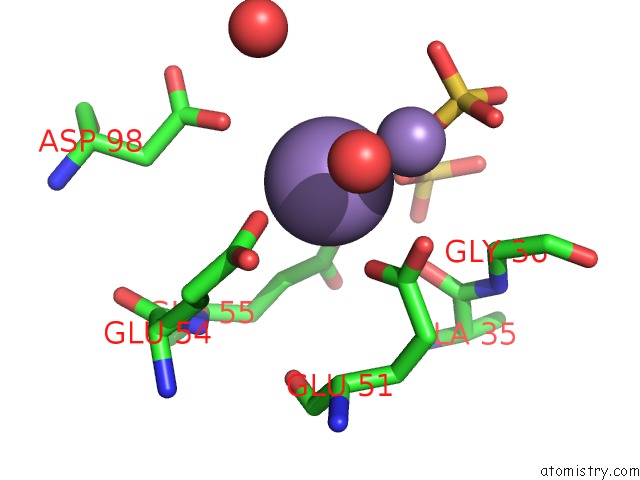
Mono view
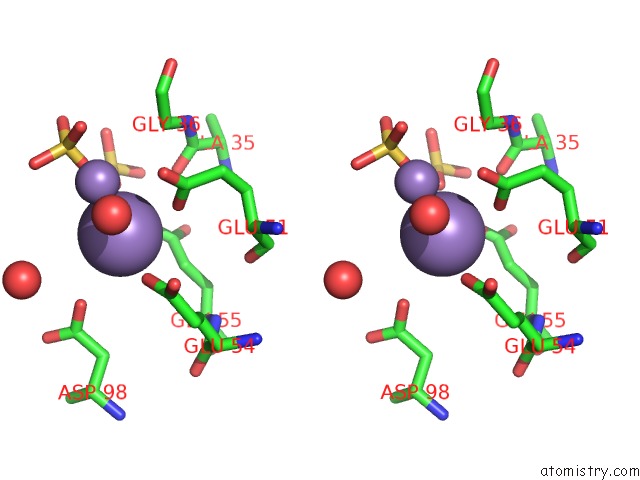
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli within 5.0Å range:
|
Manganese binding site 4 out of 22 in 4nfw
Go back to
Manganese binding site 4 out
of 22 in the Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli
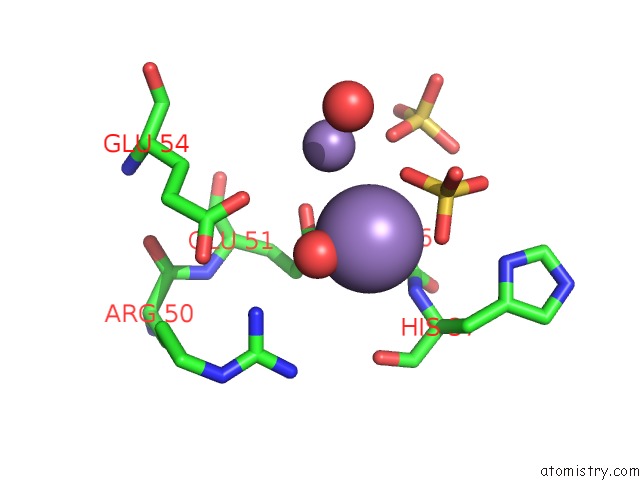
Mono view
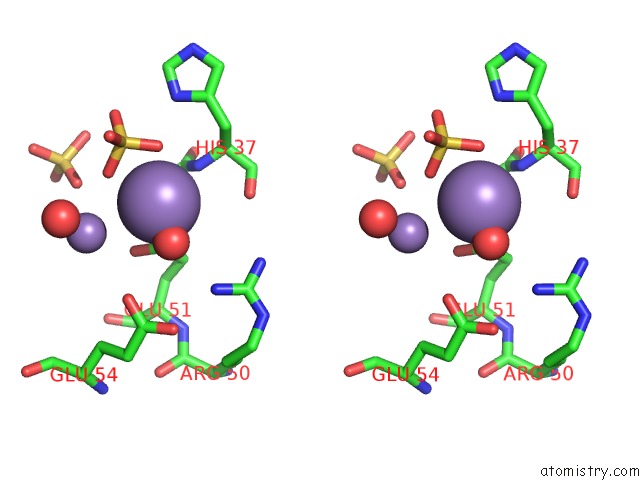
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli within 5.0Å range:
|
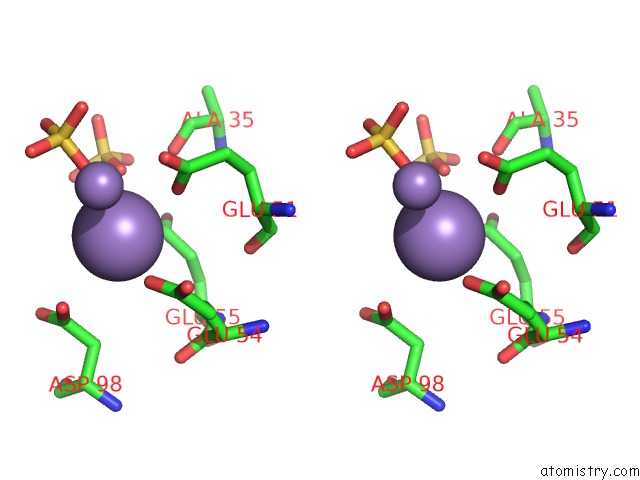
Manganese binding site 5 out of 22 in 4nfw
Go back to
Manganese binding site 5 out
of 22 in the Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli
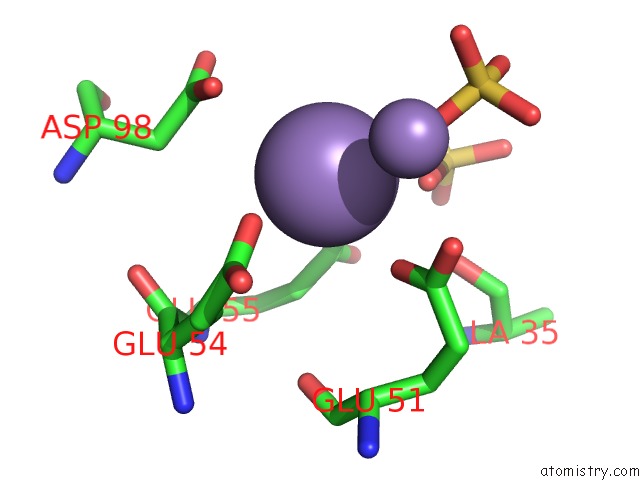
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 5 of Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli within 5.0Å range:
|
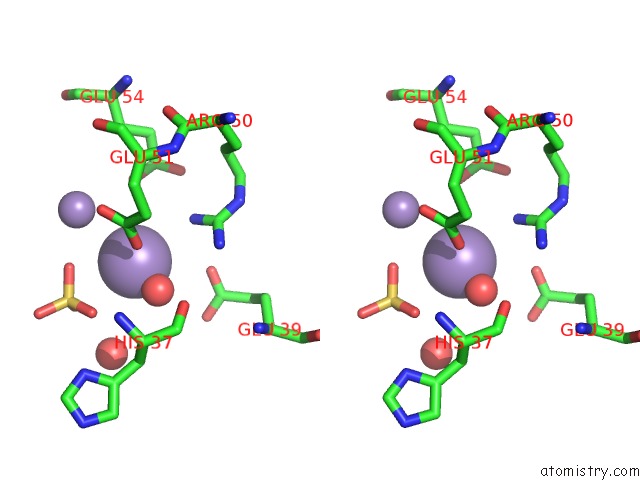
Manganese binding site 6 out of 22 in 4nfw
Go back to
Manganese binding site 6 out
of 22 in the Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli
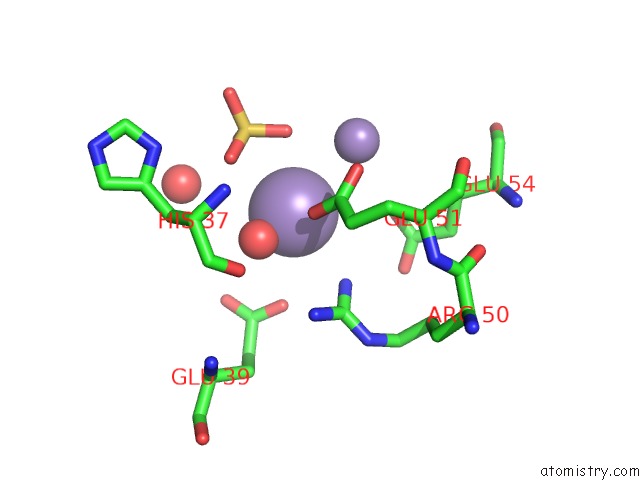
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 6 of Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli within 5.0Å range:
|
Manganese binding site 7 out of 22 in 4nfw
Go back to
Manganese binding site 7 out
of 22 in the Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli
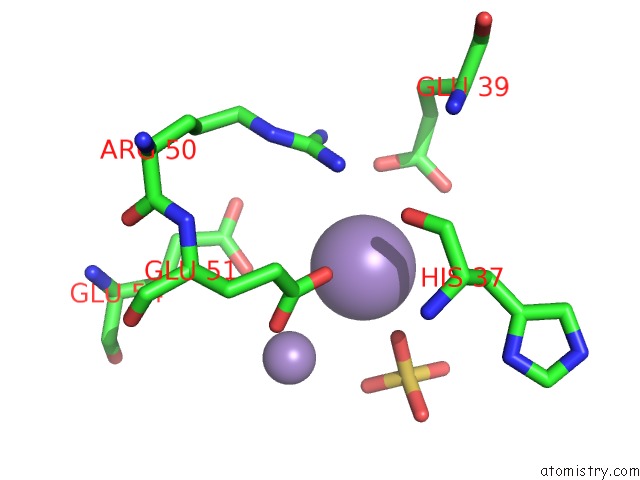
Mono view
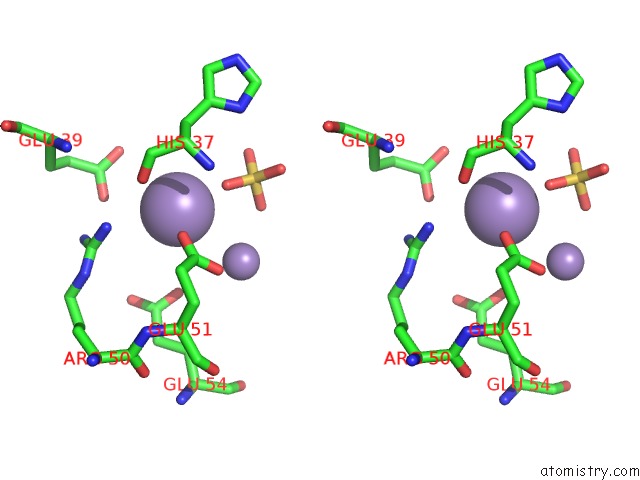
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 7 of Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli within 5.0Å range:
|
Manganese binding site 8 out of 22 in 4nfw
Go back to
Manganese binding site 8 out
of 22 in the Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli
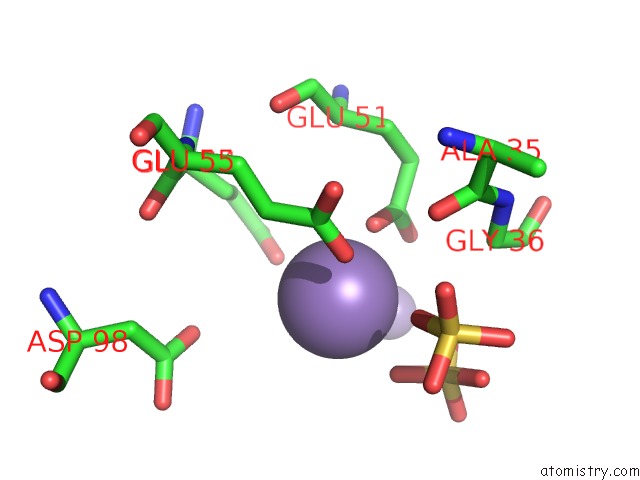
Mono view
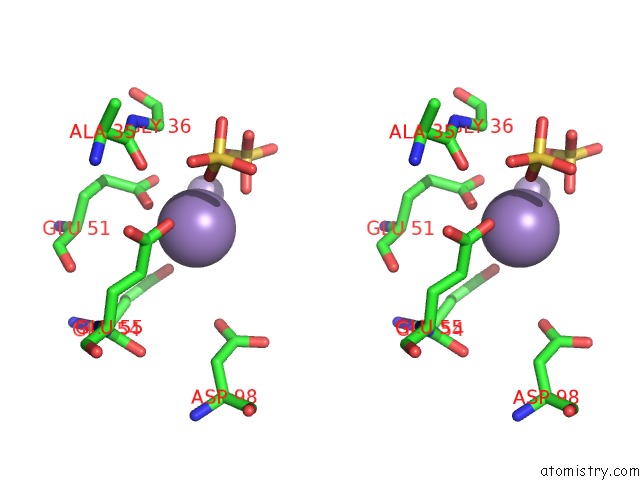
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 8 of Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli within 5.0Å range:
|
Manganese binding site 9 out of 22 in 4nfw
Go back to
Manganese binding site 9 out
of 22 in the Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli
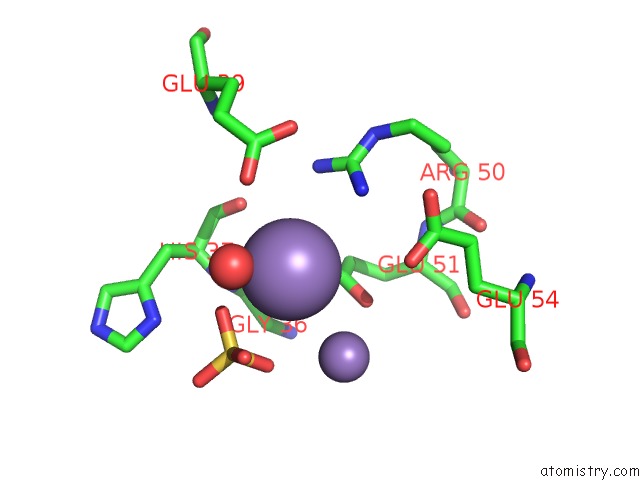
Mono view
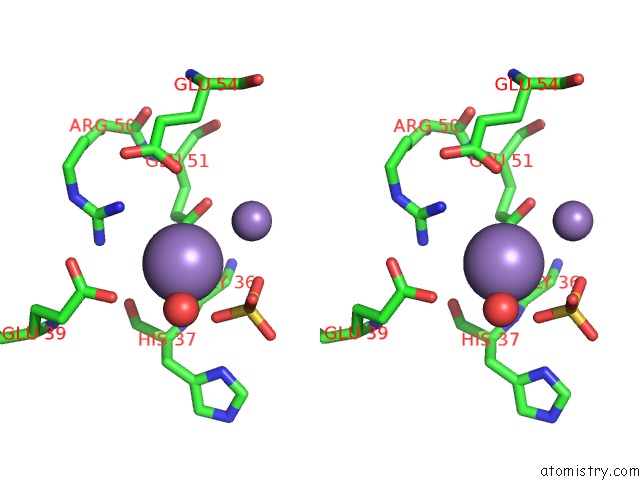
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 9 of Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli within 5.0Å range:
|
Manganese binding site 10 out of 22 in 4nfw
Go back to
Manganese binding site 10 out
of 22 in the Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli
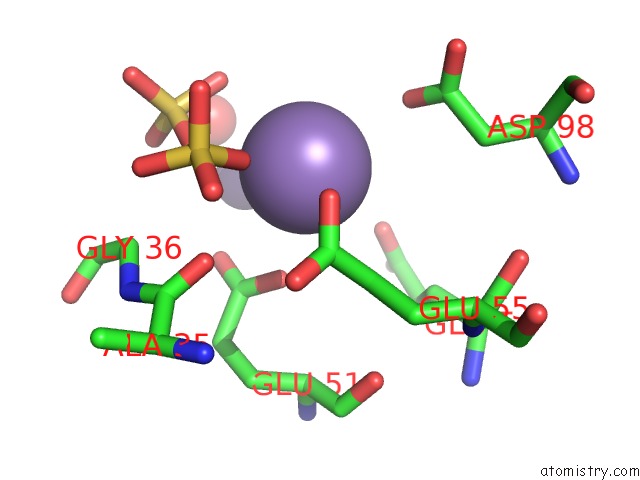
Mono view
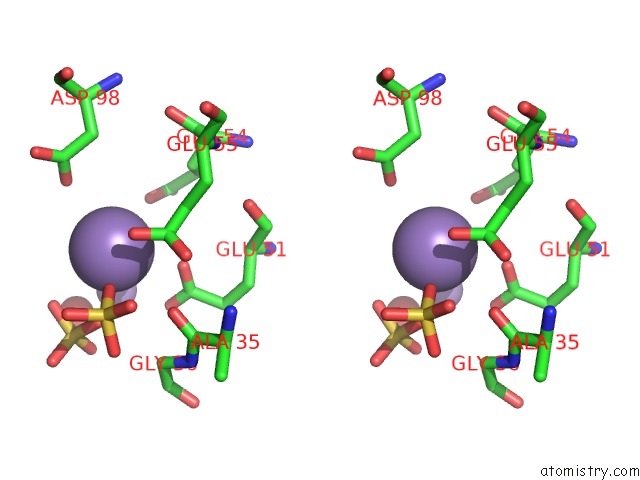
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 10 of Structure and Atypical Hydrolysis Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli within 5.0Å range:
|
Reference:
M.K.Hong,
A.J.M.Ribeiro,
J.K.Kim,
H.P.T.Ngo,
J.Kim,
C.H.Lee,
Y.J.Ahn,
P.A.Fernandes,
Q.Li,
M.J.Ramos,
L.W.Kang.
Divalent Metal Ion-Based Catalytic Mechanism of the Nudix Hydrolase ORF153 (Ymfb) From Escherichia Coli Acta Crystallogr.,Sect.D V. 70 1297 2014.
ISSN: ISSN 0907-4449
DOI: 10.1107/S1399004714002570
Page generated: Sat Aug 16 14:46:49 2025
ISSN: ISSN 0907-4449
DOI: 10.1107/S1399004714002570
Last articles
Ni in 5Q9CNi in 5Q9B
Ni in 5Q9A
Ni in 5Q9D
Ni in 5Q99
Ni in 5Q96
Ni in 5Q97
Ni in 5Q98
Ni in 5Q93
Ni in 5Q94