Manganese »
PDB 4dky-4ee1 »
4edr »
Manganese in PDB 4edr: The Structure of the S. Aureus Dnag Rna Polymerase Domain Bound to Utp and Manganese
Protein crystallography data
The structure of The Structure of the S. Aureus Dnag Rna Polymerase Domain Bound to Utp and Manganese, PDB code: 4edr
was solved by
R.U.Rymer,
F.A.Solorio,
C.Chu,
J.E.Corn,
J.D.Wang,
J.M.Berger,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 37.04 / 2.01 |
| Space group | P 61 |
| Cell size a, b, c (Å), α, β, γ (°) | 150.582, 150.582, 38.633, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 17.2 / 20.5 |
Manganese Binding Sites:
The binding sites of Manganese atom in the The Structure of the S. Aureus Dnag Rna Polymerase Domain Bound to Utp and Manganese
(pdb code 4edr). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 3 binding sites of Manganese where determined in the The Structure of the S. Aureus Dnag Rna Polymerase Domain Bound to Utp and Manganese, PDB code: 4edr:
Jump to Manganese binding site number: 1; 2; 3;
In total 3 binding sites of Manganese where determined in the The Structure of the S. Aureus Dnag Rna Polymerase Domain Bound to Utp and Manganese, PDB code: 4edr:
Jump to Manganese binding site number: 1; 2; 3;
Manganese binding site 1 out of 3 in 4edr
Go back to
Manganese binding site 1 out
of 3 in the The Structure of the S. Aureus Dnag Rna Polymerase Domain Bound to Utp and Manganese
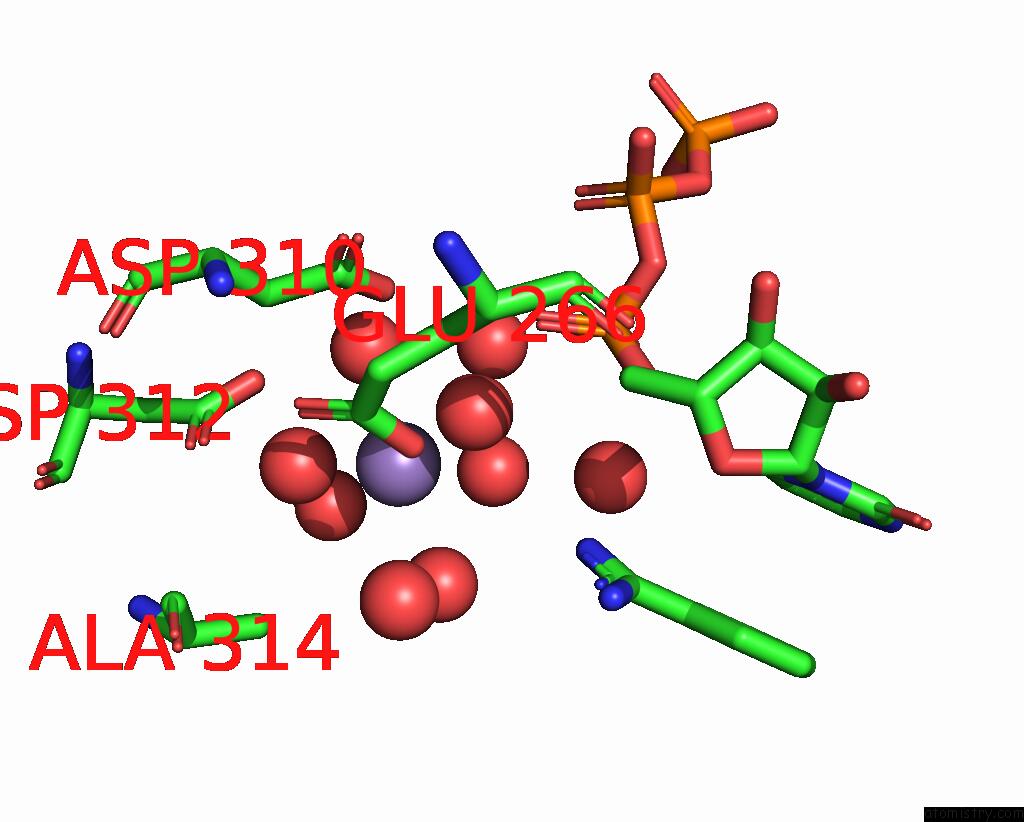
Mono view
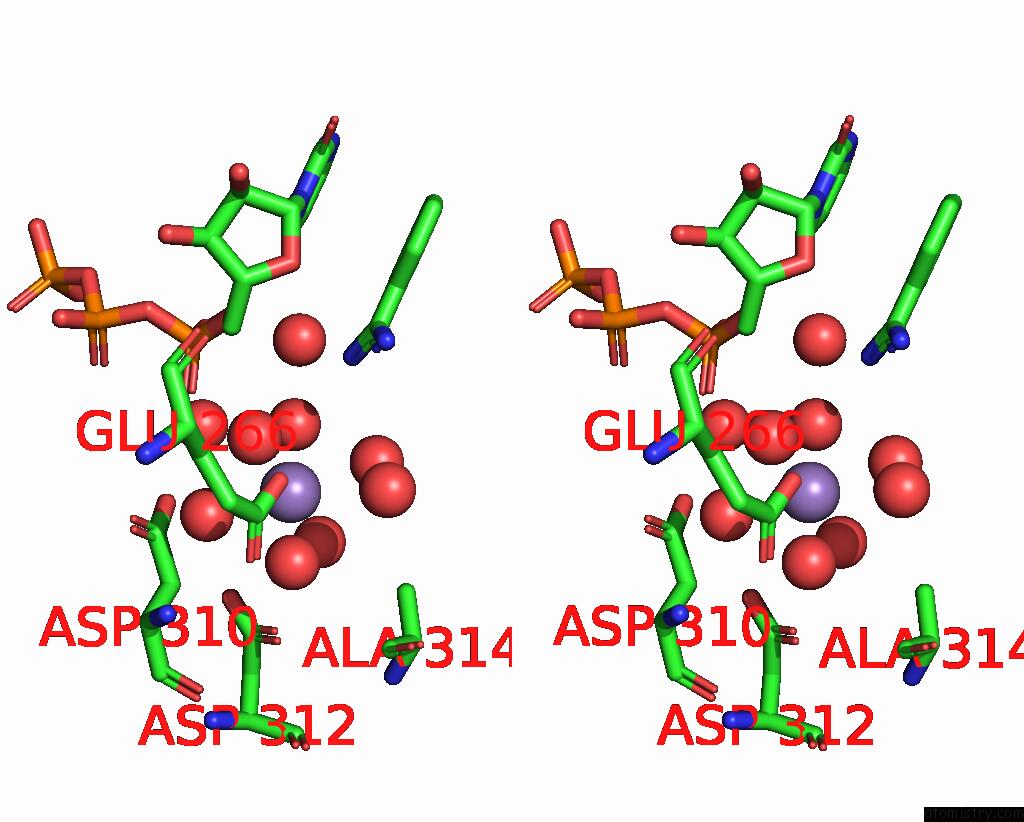
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of The Structure of the S. Aureus Dnag Rna Polymerase Domain Bound to Utp and Manganese within 5.0Å range:
|
Manganese binding site 2 out of 3 in 4edr
Go back to
Manganese binding site 2 out
of 3 in the The Structure of the S. Aureus Dnag Rna Polymerase Domain Bound to Utp and Manganese
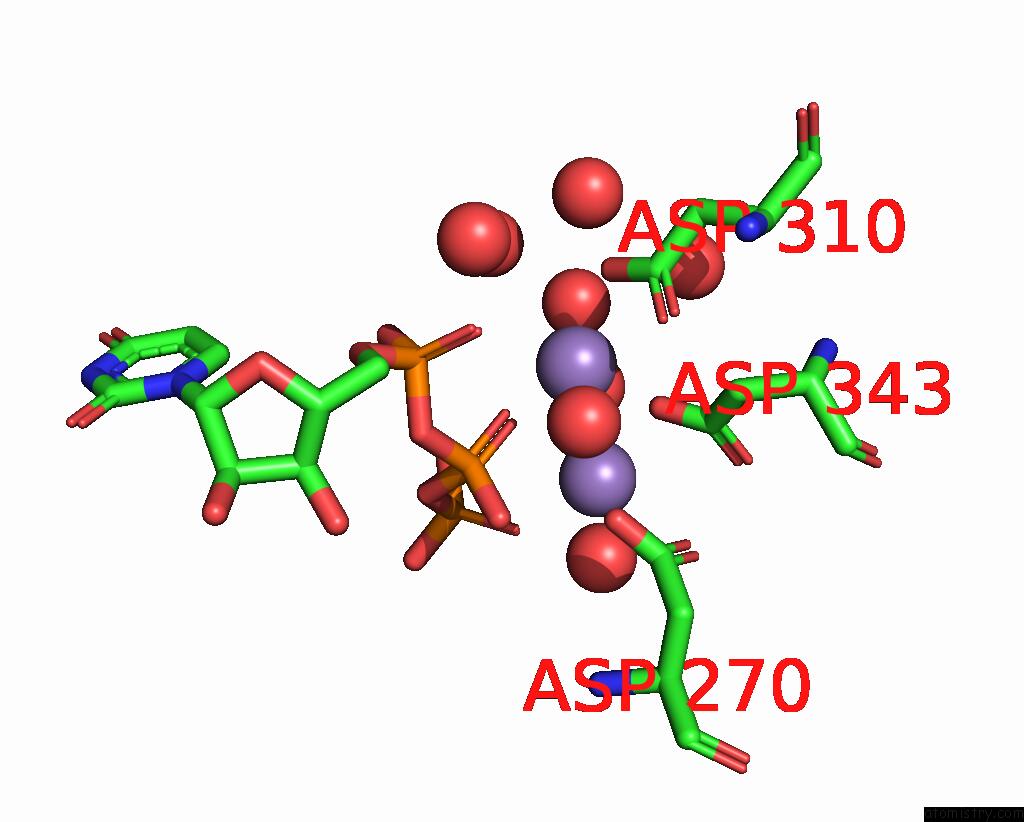
Mono view
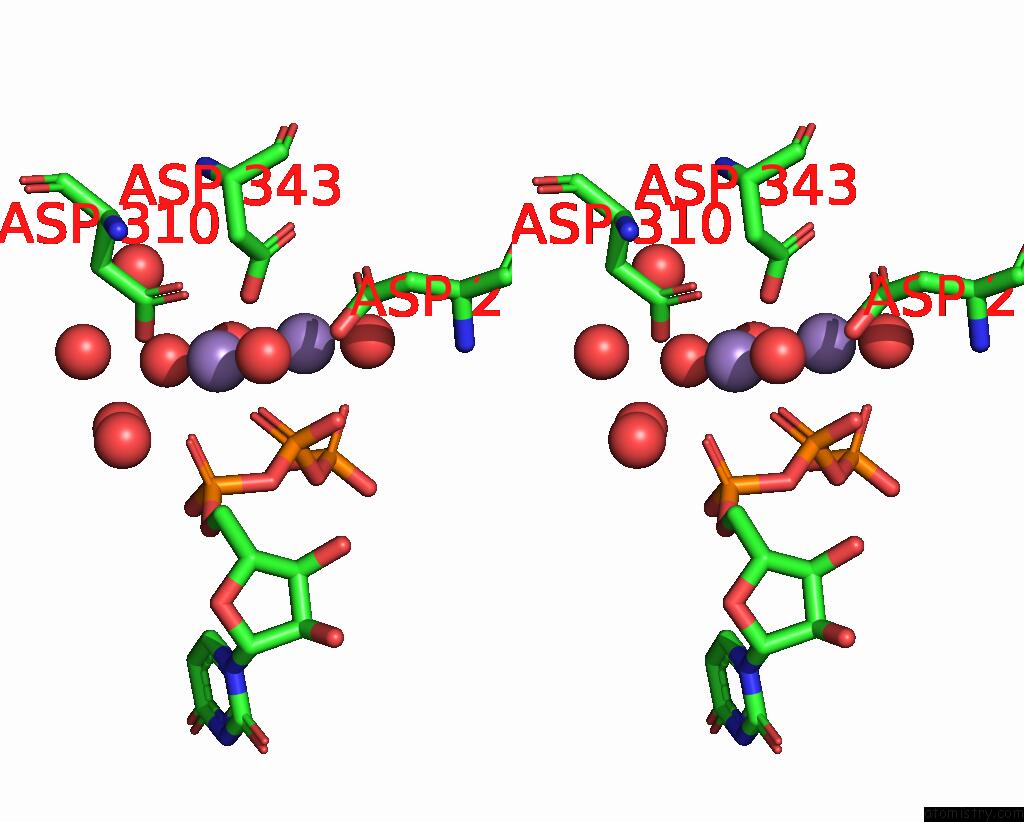
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of The Structure of the S. Aureus Dnag Rna Polymerase Domain Bound to Utp and Manganese within 5.0Å range:
|
Manganese binding site 3 out of 3 in 4edr
Go back to
Manganese binding site 3 out
of 3 in the The Structure of the S. Aureus Dnag Rna Polymerase Domain Bound to Utp and Manganese
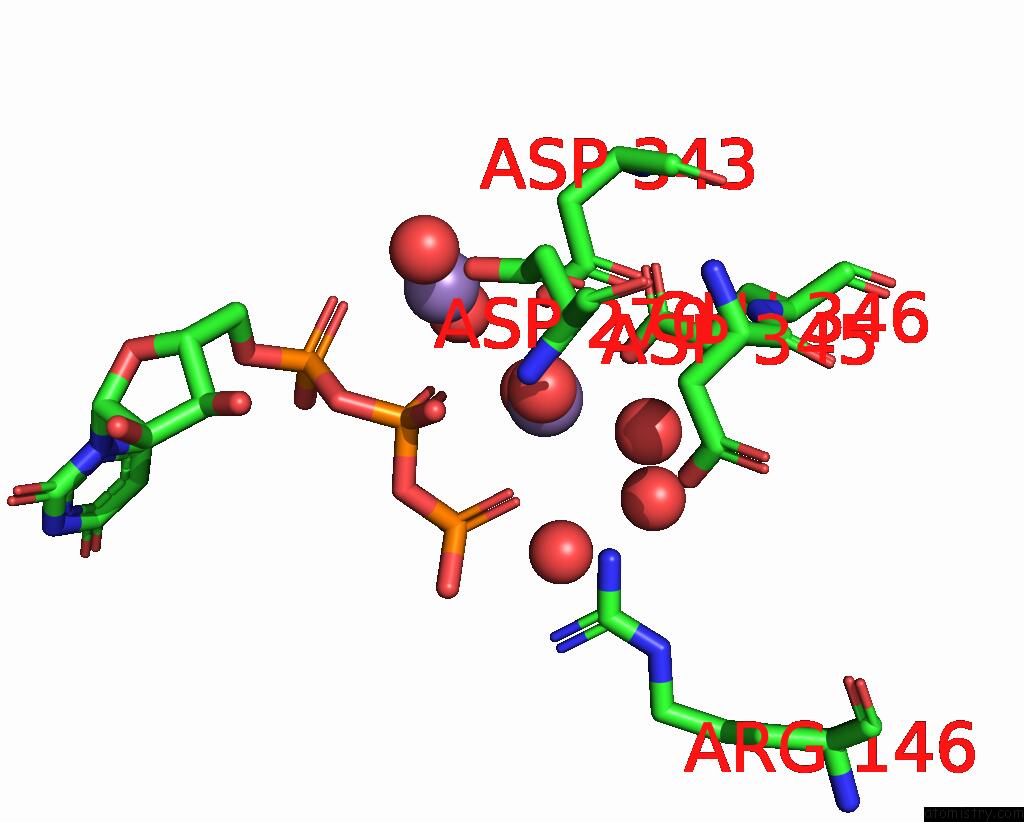
Mono view
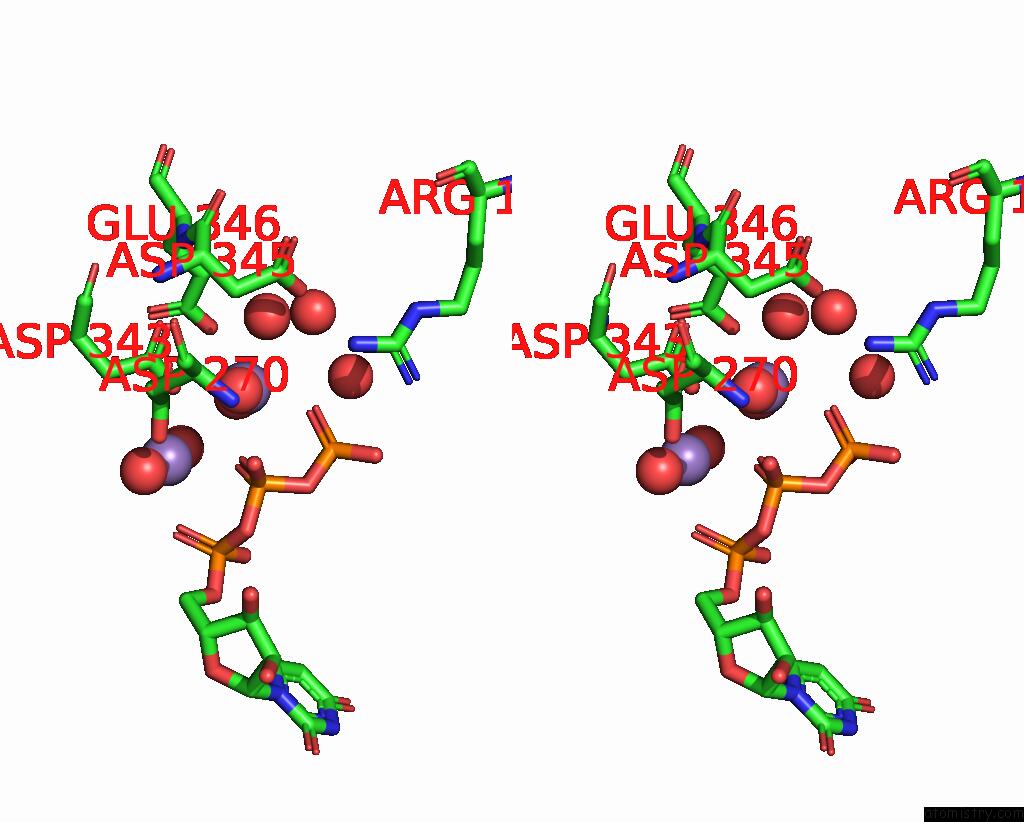
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of The Structure of the S. Aureus Dnag Rna Polymerase Domain Bound to Utp and Manganese within 5.0Å range:
|
Reference:
R.U.Rymer,
F.A.Solorio,
A.K.Tehranchi,
C.Chu,
J.E.Corn,
J.L.Keck,
J.D.Wang,
J.M.Berger.
Binding Mechanism of Metal-Ntp Substrates and Stringent-Response Alarmones to Bacterial Dnag-Type Primases. Structure V. 20 1478 2012.
ISSN: ISSN 0969-2126
PubMed: 22795082
DOI: 10.1016/J.STR.2012.05.017
Page generated: Sat Aug 16 13:53:44 2025
ISSN: ISSN 0969-2126
PubMed: 22795082
DOI: 10.1016/J.STR.2012.05.017
Last articles
Na in 3DIQNa in 3DIO
Na in 3DIM
Na in 3DIG
Na in 3DH4
Na in 3DHK
Na in 3CXC
Na in 3DDK
Na in 3DFH
Na in 3DEB