Manganese »
PDB 4dky-4ee1 »
4e2s »
Manganese in PDB 4e2s: Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine
Protein crystallography data
The structure of Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine, PDB code: 4e2s
was solved by
I.Shin,
S.Rhee,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.59 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 93.098, 174.894, 154.300, 90.00, 99.26, 90.00 |
| R / Rfree (%) | 20.9 / 27.1 |
Manganese Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 16;Binding sites:
The binding sites of Manganese atom in the Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine (pdb code 4e2s). This binding sites where shown within 5.0 Angstroms radius around Manganese atom.In total 16 binding sites of Manganese where determined in the Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine, PDB code: 4e2s:
Jump to Manganese binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
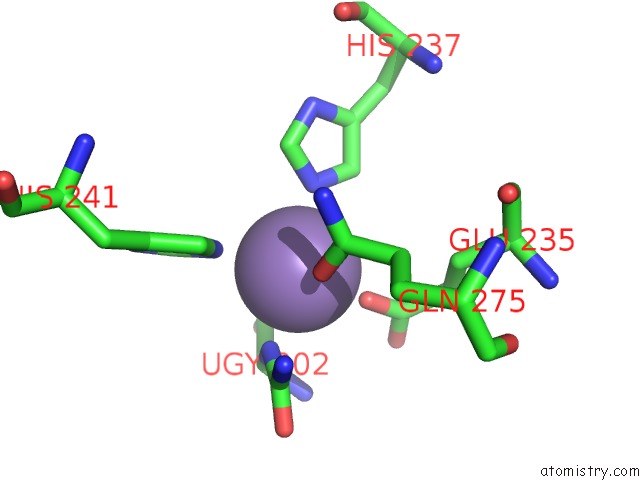
Manganese binding site 1 out of 16 in 4e2s
Go back to
Manganese binding site 1 out
of 16 in the Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine
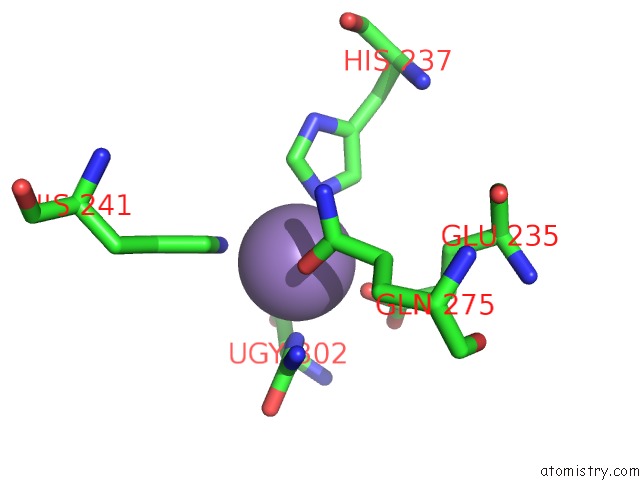
Mono view
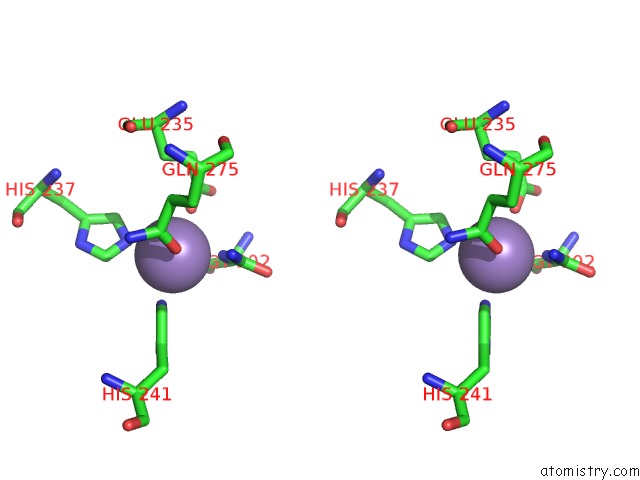
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine within 5.0Å range:
|
Manganese binding site 2 out of 16 in 4e2s
Go back to
Manganese binding site 2 out
of 16 in the Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine
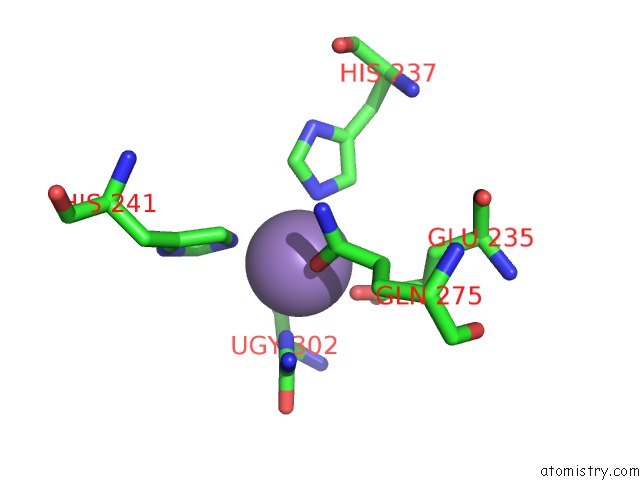
Mono view
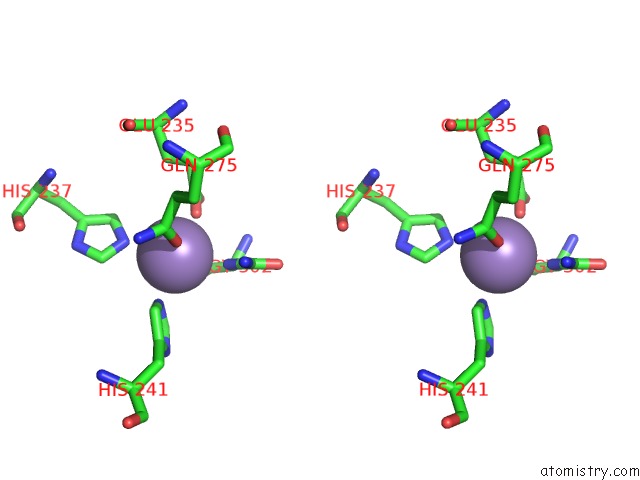
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine within 5.0Å range:
|
Manganese binding site 3 out of 16 in 4e2s
Go back to
Manganese binding site 3 out
of 16 in the Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine
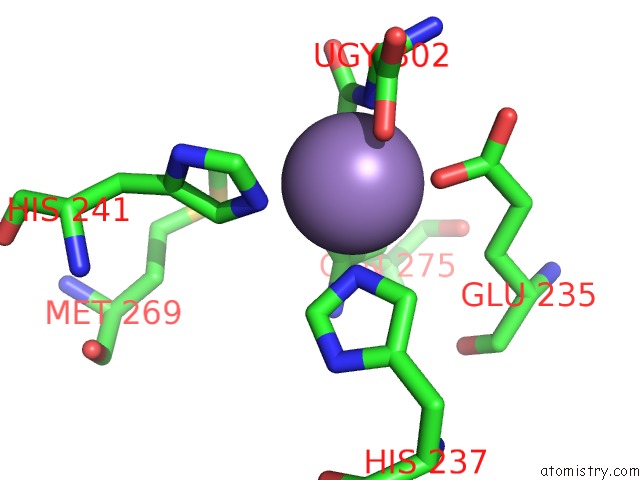
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine within 5.0Å range:
|
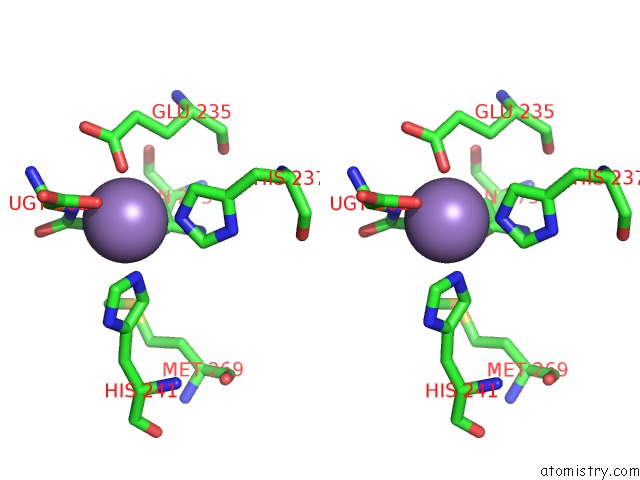
Manganese binding site 4 out of 16 in 4e2s
Go back to
Manganese binding site 4 out
of 16 in the Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine within 5.0Å range:
|
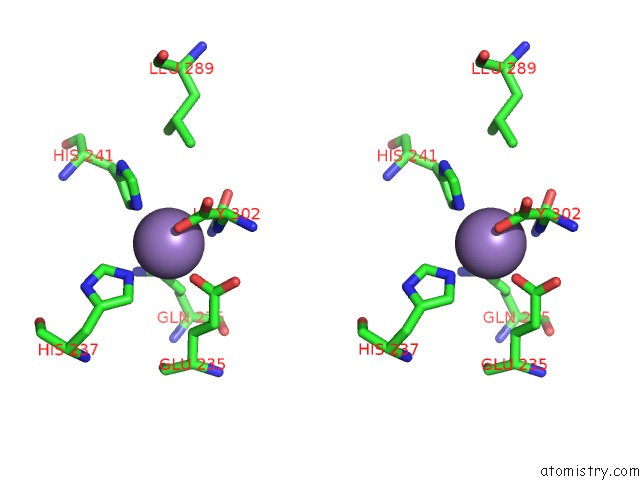
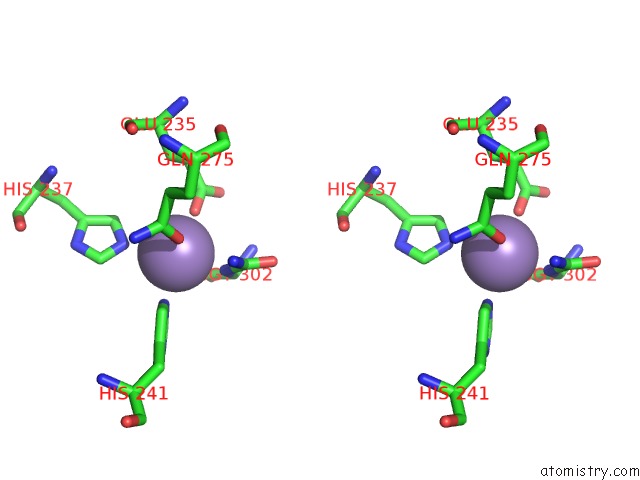
Manganese binding site 5 out of 16 in 4e2s
Go back to
Manganese binding site 5 out
of 16 in the Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 5 of Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine within 5.0Å range:
|
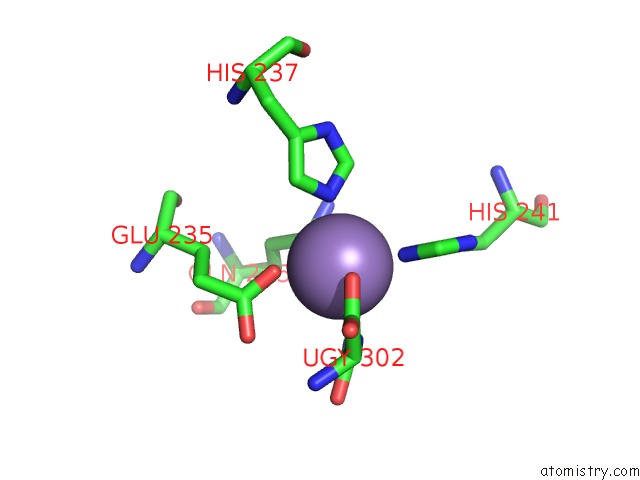
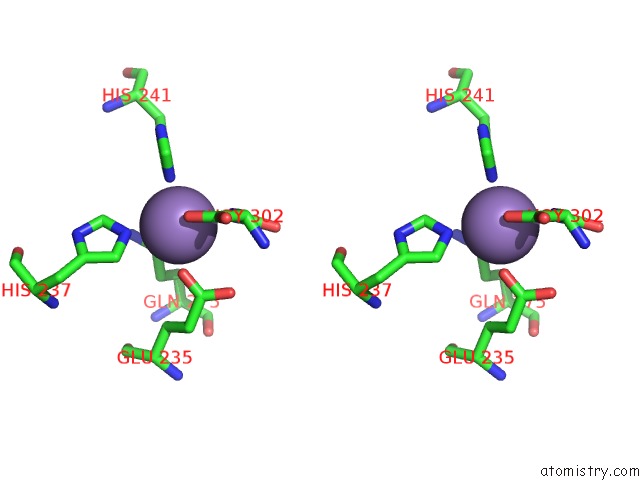
Manganese binding site 6 out of 16 in 4e2s
Go back to
Manganese binding site 6 out
of 16 in the Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 6 of Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine within 5.0Å range:
|
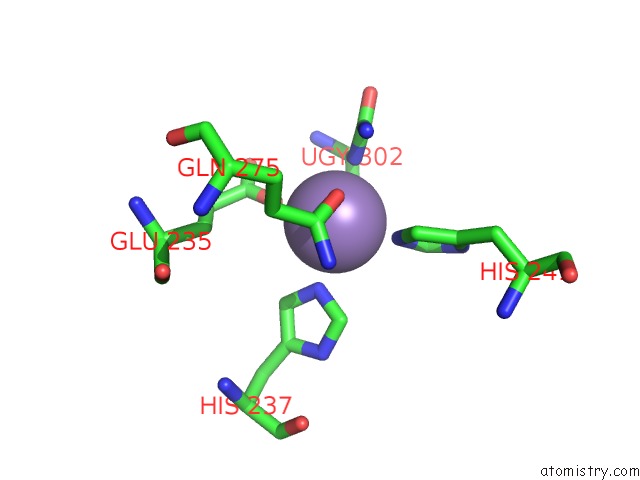
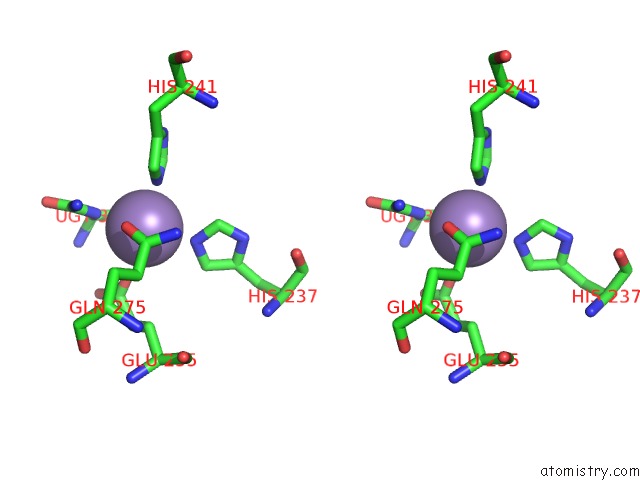
Manganese binding site 7 out of 16 in 4e2s
Go back to
Manganese binding site 7 out
of 16 in the Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 7 of Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine within 5.0Å range:
|
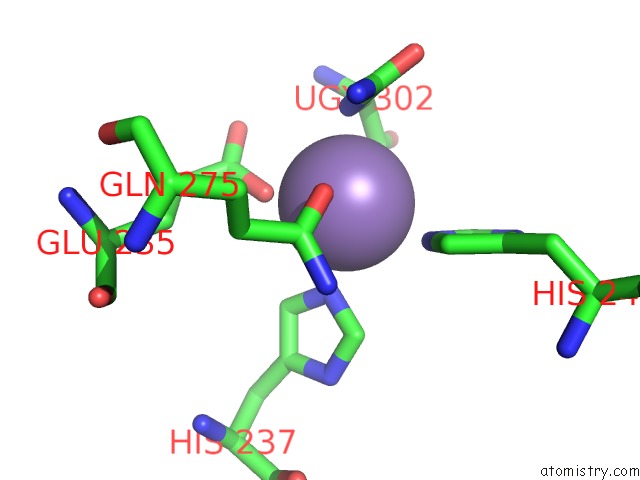
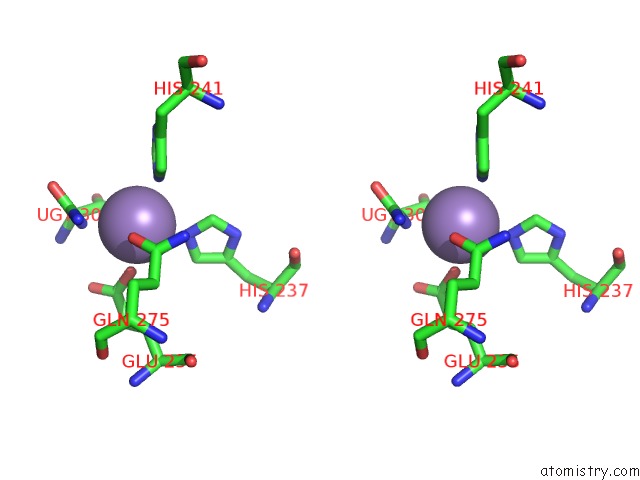
Manganese binding site 8 out of 16 in 4e2s
Go back to
Manganese binding site 8 out
of 16 in the Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 8 of Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine within 5.0Å range:
|
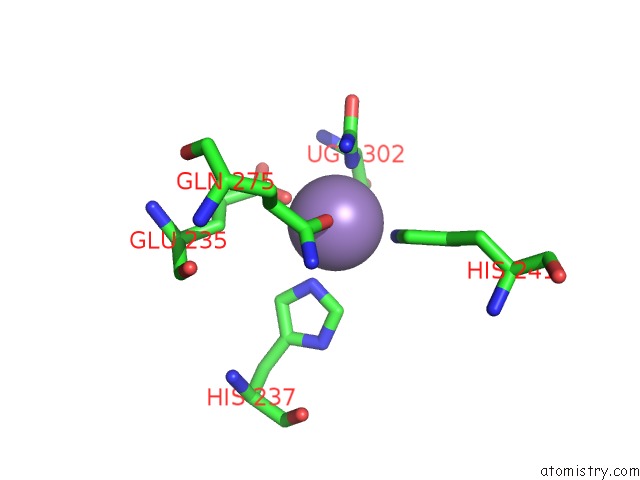
Manganese binding site 9 out of 16 in 4e2s
Go back to
Manganese binding site 9 out
of 16 in the Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine
Mono view
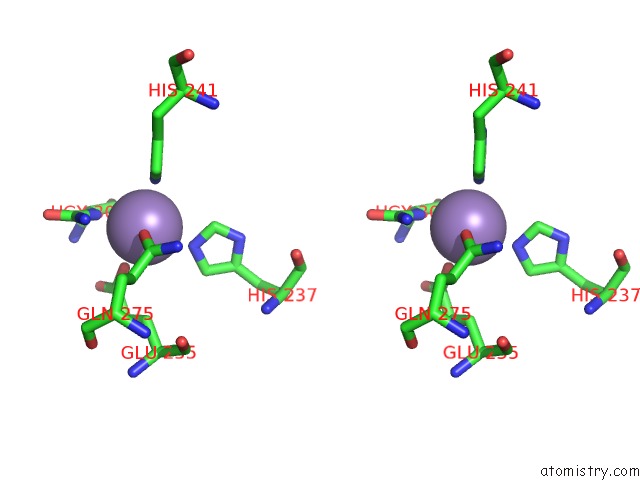
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 9 of Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine within 5.0Å range:
|
Manganese binding site 10 out of 16 in 4e2s
Go back to
Manganese binding site 10 out
of 16 in the Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine
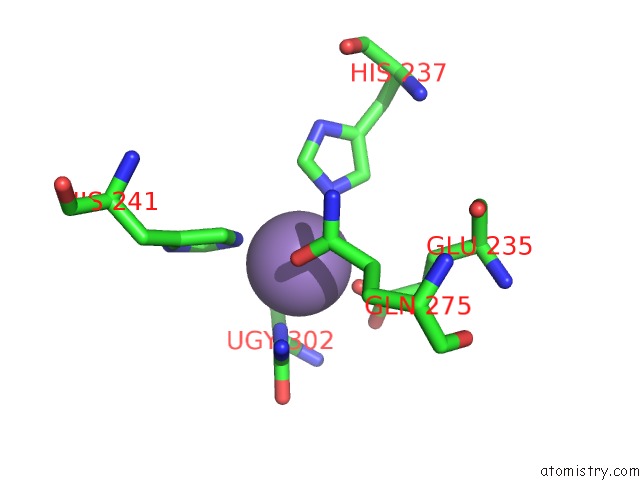
Mono view
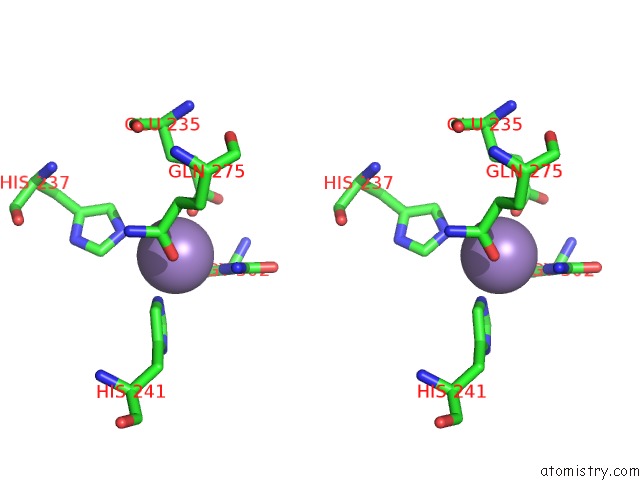
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 10 of Crystal Structure of (S)-Ureidoglycine Aminohydrolase From Arabidopsis Thaliana in Complex with Its Substrate, (S)-Ureidoglycine within 5.0Å range:
|
Reference:
I.Shin,
R.Percudani,
S.Rhee.
Structural and Functional Insights Into (S)-Ureidoglycine Aminohydrolase, Key Enzyme of Purine Catabolism in Arabidopsis Thaliana J.Biol.Chem. V. 287 18796 2012.
ISSN: ISSN 0021-9258
PubMed: 22493446
DOI: 10.1074/JBC.M111.331819
Page generated: Sat Aug 16 13:47:24 2025
ISSN: ISSN 0021-9258
PubMed: 22493446
DOI: 10.1074/JBC.M111.331819
Last articles
Mn in 9MBIMn in 9MBH
Mn in 9L56
Mn in 9KOQ
Mn in 9KOJ
Mn in 9KQC
Mn in 9JOA
Mn in 9JOB
Mn in 9JHN
Mn in 9JL7