Manganese »
PDB 3n39-3orm »
3nvt »
Manganese in PDB 3nvt: 1.95 Angstrom Crystal Structure of A Bifunctional 3-Deoxy-7- Phosphoheptulonate Synthase/Chorismate Mutase (Aroa) From Listeria Monocytogenes Egd-E
Enzymatic activity of 1.95 Angstrom Crystal Structure of A Bifunctional 3-Deoxy-7- Phosphoheptulonate Synthase/Chorismate Mutase (Aroa) From Listeria Monocytogenes Egd-E
All present enzymatic activity of 1.95 Angstrom Crystal Structure of A Bifunctional 3-Deoxy-7- Phosphoheptulonate Synthase/Chorismate Mutase (Aroa) From Listeria Monocytogenes Egd-E:
2.5.1.54; 5.4.99.5;
2.5.1.54; 5.4.99.5;
Protein crystallography data
The structure of 1.95 Angstrom Crystal Structure of A Bifunctional 3-Deoxy-7- Phosphoheptulonate Synthase/Chorismate Mutase (Aroa) From Listeria Monocytogenes Egd-E, PDB code: 3nvt
was solved by
A.S.Halavaty,
S.H.Light,
G.Minasov,
L.Shuvalova,
K.Kwon,
W.F.Anderson,
Center For Structural Genomics Of Infectious Diseases (Csgid),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.04 / 1.95 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 111.570, 111.787, 81.049, 90.00, 127.63, 90.00 |
| R / Rfree (%) | 15.4 / 19.8 |
Manganese Binding Sites:
The binding sites of Manganese atom in the 1.95 Angstrom Crystal Structure of A Bifunctional 3-Deoxy-7- Phosphoheptulonate Synthase/Chorismate Mutase (Aroa) From Listeria Monocytogenes Egd-E
(pdb code 3nvt). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 4 binding sites of Manganese where determined in the 1.95 Angstrom Crystal Structure of A Bifunctional 3-Deoxy-7- Phosphoheptulonate Synthase/Chorismate Mutase (Aroa) From Listeria Monocytogenes Egd-E, PDB code: 3nvt:
Jump to Manganese binding site number: 1; 2; 3; 4;
In total 4 binding sites of Manganese where determined in the 1.95 Angstrom Crystal Structure of A Bifunctional 3-Deoxy-7- Phosphoheptulonate Synthase/Chorismate Mutase (Aroa) From Listeria Monocytogenes Egd-E, PDB code: 3nvt:
Jump to Manganese binding site number: 1; 2; 3; 4;
Manganese binding site 1 out of 4 in 3nvt
Go back to
Manganese binding site 1 out
of 4 in the 1.95 Angstrom Crystal Structure of A Bifunctional 3-Deoxy-7- Phosphoheptulonate Synthase/Chorismate Mutase (Aroa) From Listeria Monocytogenes Egd-E
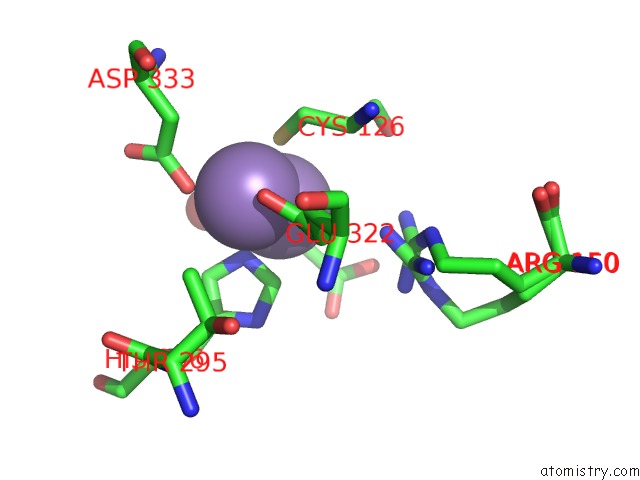
Mono view
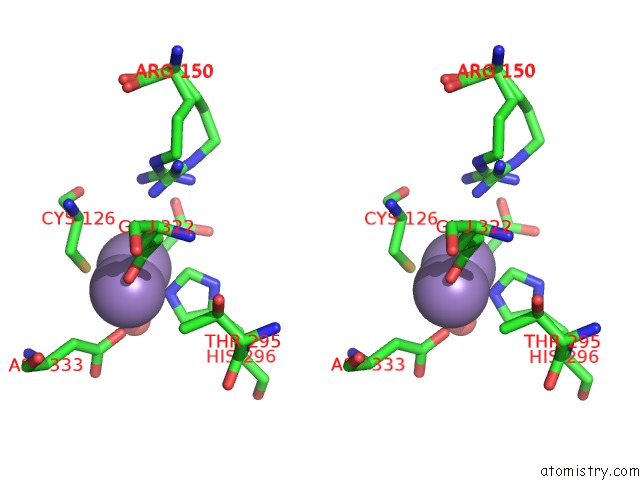
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of 1.95 Angstrom Crystal Structure of A Bifunctional 3-Deoxy-7- Phosphoheptulonate Synthase/Chorismate Mutase (Aroa) From Listeria Monocytogenes Egd-E within 5.0Å range:
|
Manganese binding site 2 out of 4 in 3nvt
Go back to
Manganese binding site 2 out
of 4 in the 1.95 Angstrom Crystal Structure of A Bifunctional 3-Deoxy-7- Phosphoheptulonate Synthase/Chorismate Mutase (Aroa) From Listeria Monocytogenes Egd-E
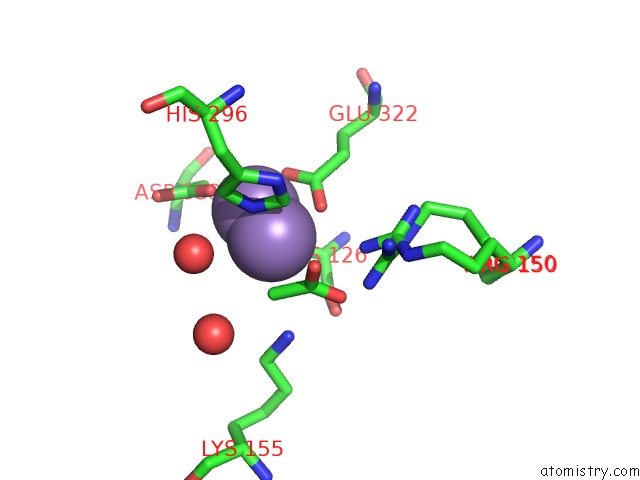
Mono view
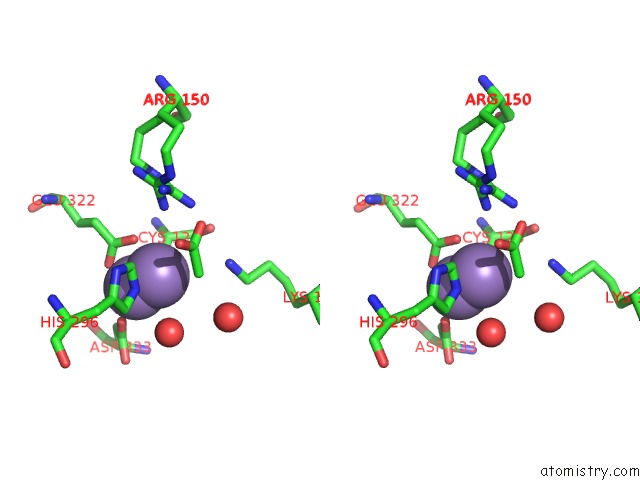
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of 1.95 Angstrom Crystal Structure of A Bifunctional 3-Deoxy-7- Phosphoheptulonate Synthase/Chorismate Mutase (Aroa) From Listeria Monocytogenes Egd-E within 5.0Å range:
|
Manganese binding site 3 out of 4 in 3nvt
Go back to
Manganese binding site 3 out
of 4 in the 1.95 Angstrom Crystal Structure of A Bifunctional 3-Deoxy-7- Phosphoheptulonate Synthase/Chorismate Mutase (Aroa) From Listeria Monocytogenes Egd-E
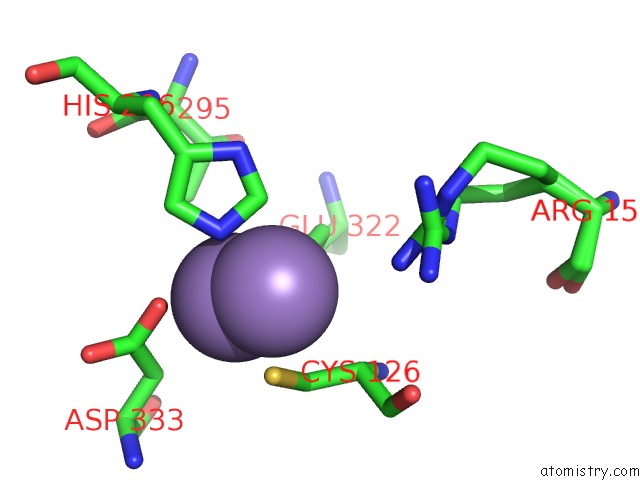
Mono view
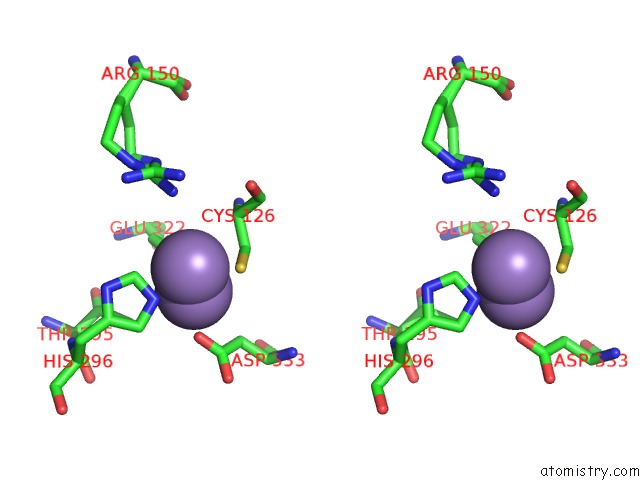
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of 1.95 Angstrom Crystal Structure of A Bifunctional 3-Deoxy-7- Phosphoheptulonate Synthase/Chorismate Mutase (Aroa) From Listeria Monocytogenes Egd-E within 5.0Å range:
|
Manganese binding site 4 out of 4 in 3nvt
Go back to
Manganese binding site 4 out
of 4 in the 1.95 Angstrom Crystal Structure of A Bifunctional 3-Deoxy-7- Phosphoheptulonate Synthase/Chorismate Mutase (Aroa) From Listeria Monocytogenes Egd-E
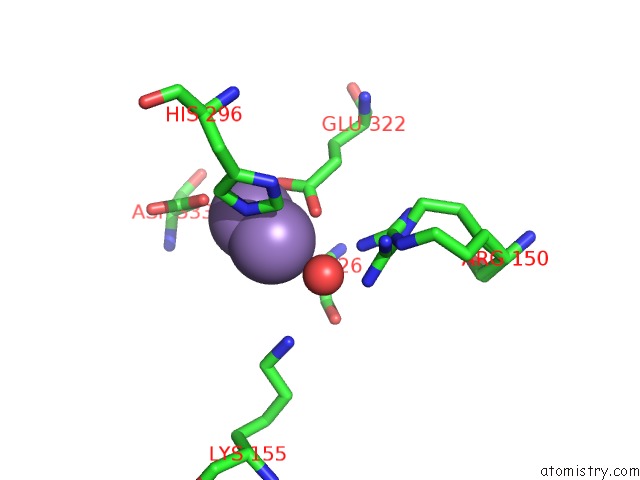
Mono view
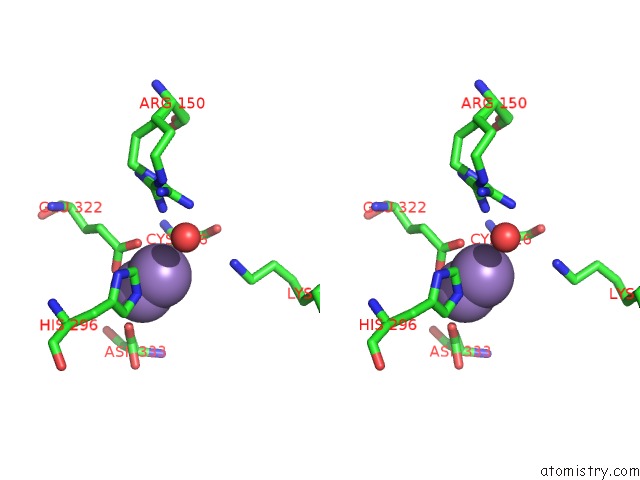
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of 1.95 Angstrom Crystal Structure of A Bifunctional 3-Deoxy-7- Phosphoheptulonate Synthase/Chorismate Mutase (Aroa) From Listeria Monocytogenes Egd-E within 5.0Å range:
|
Reference:
S.H.Light,
A.S.Halavaty,
G.Minasov,
L.Shuvalova,
W.F.Anderson.
Structural Analysis of A 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase with An N-Terminal Chorismate Mutase-Like Regulatory Domain. Protein Sci. V. 21 887 2012.
ISSN: ISSN 0961-8368
PubMed: 22505283
DOI: 10.1002/PRO.2075
Page generated: Sat Oct 5 17:21:27 2024
ISSN: ISSN 0961-8368
PubMed: 22505283
DOI: 10.1002/PRO.2075
Last articles
Cl in 5VJOCl in 5VJA
Cl in 5VFB
Cl in 5VJ2
Cl in 5VIV
Cl in 5VJ1
Cl in 5VIS
Cl in 5VIF
Cl in 5VH6
Cl in 5VIE