Manganese »
PDB 3g0z-3hvq »
3gx6 »
Manganese in PDB 3gx6: Crystal Structure of the T. Tengcongensis Sam-I Riboswitch Variant U34C/A94G Bound with Sam in Manganese Chloride
Protein crystallography data
The structure of Crystal Structure of the T. Tengcongensis Sam-I Riboswitch Variant U34C/A94G Bound with Sam in Manganese Chloride, PDB code: 3gx6
was solved by
R.K.Montange,
R.T.Batey,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 18.47 / 2.80 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 62.390, 62.390, 157.844, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 26 / 32.7 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Crystal Structure of the T. Tengcongensis Sam-I Riboswitch Variant U34C/A94G Bound with Sam in Manganese Chloride
(pdb code 3gx6). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 5 binding sites of Manganese where determined in the Crystal Structure of the T. Tengcongensis Sam-I Riboswitch Variant U34C/A94G Bound with Sam in Manganese Chloride, PDB code: 3gx6:
Jump to Manganese binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Manganese where determined in the Crystal Structure of the T. Tengcongensis Sam-I Riboswitch Variant U34C/A94G Bound with Sam in Manganese Chloride, PDB code: 3gx6:
Jump to Manganese binding site number: 1; 2; 3; 4; 5;
Manganese binding site 1 out of 5 in 3gx6
Go back to
Manganese binding site 1 out
of 5 in the Crystal Structure of the T. Tengcongensis Sam-I Riboswitch Variant U34C/A94G Bound with Sam in Manganese Chloride
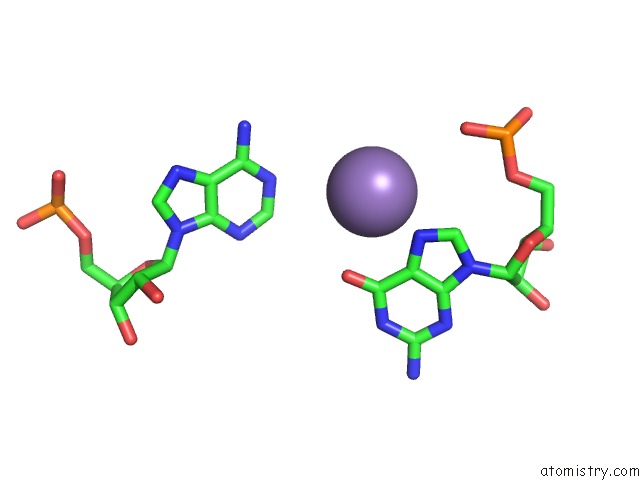
Mono view
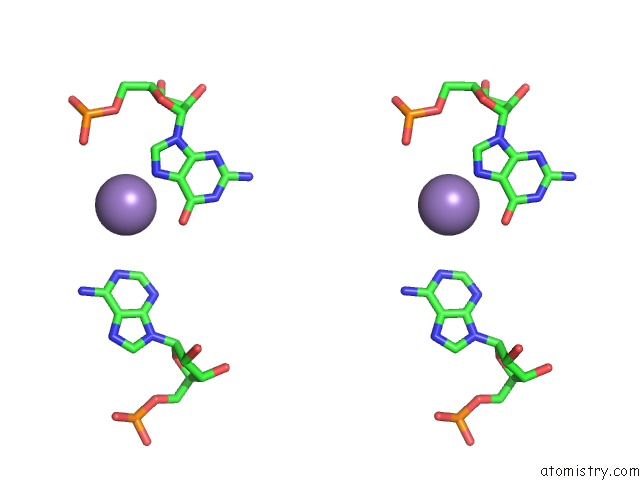
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Crystal Structure of the T. Tengcongensis Sam-I Riboswitch Variant U34C/A94G Bound with Sam in Manganese Chloride within 5.0Å range:
|
Manganese binding site 2 out of 5 in 3gx6
Go back to
Manganese binding site 2 out
of 5 in the Crystal Structure of the T. Tengcongensis Sam-I Riboswitch Variant U34C/A94G Bound with Sam in Manganese Chloride
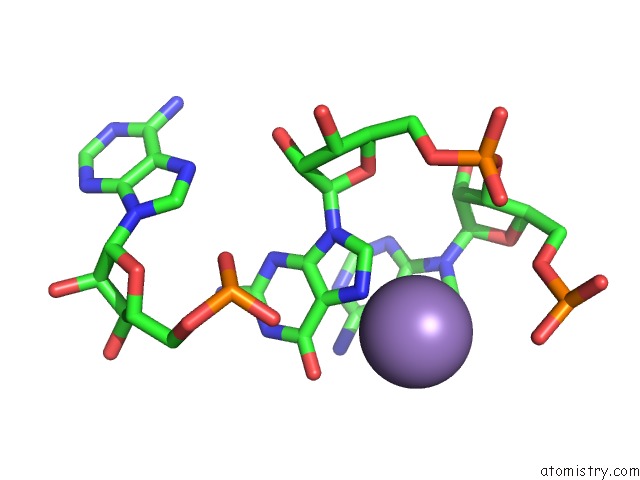
Mono view
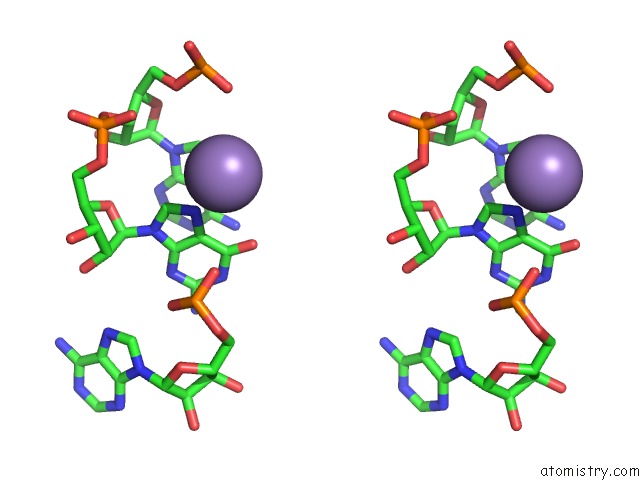
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Crystal Structure of the T. Tengcongensis Sam-I Riboswitch Variant U34C/A94G Bound with Sam in Manganese Chloride within 5.0Å range:
|
Manganese binding site 3 out of 5 in 3gx6
Go back to
Manganese binding site 3 out
of 5 in the Crystal Structure of the T. Tengcongensis Sam-I Riboswitch Variant U34C/A94G Bound with Sam in Manganese Chloride
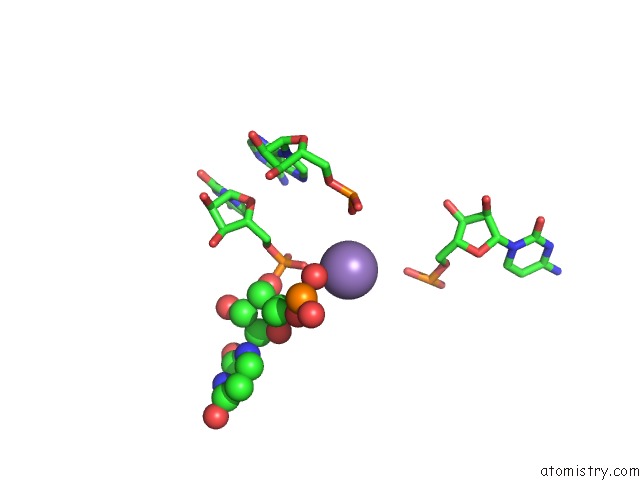
Mono view
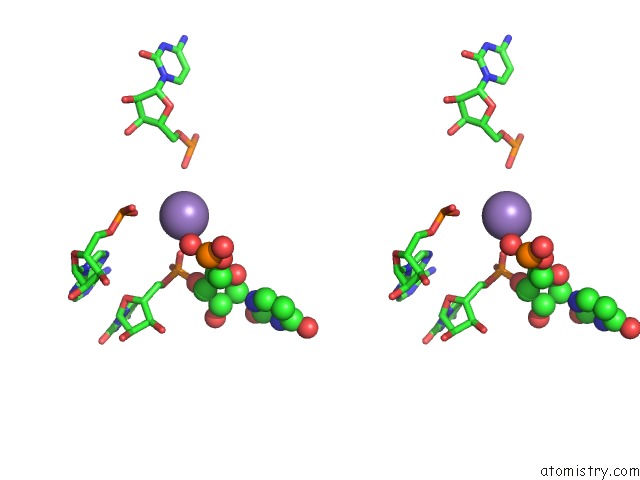
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Crystal Structure of the T. Tengcongensis Sam-I Riboswitch Variant U34C/A94G Bound with Sam in Manganese Chloride within 5.0Å range:
|
Manganese binding site 4 out of 5 in 3gx6
Go back to
Manganese binding site 4 out
of 5 in the Crystal Structure of the T. Tengcongensis Sam-I Riboswitch Variant U34C/A94G Bound with Sam in Manganese Chloride
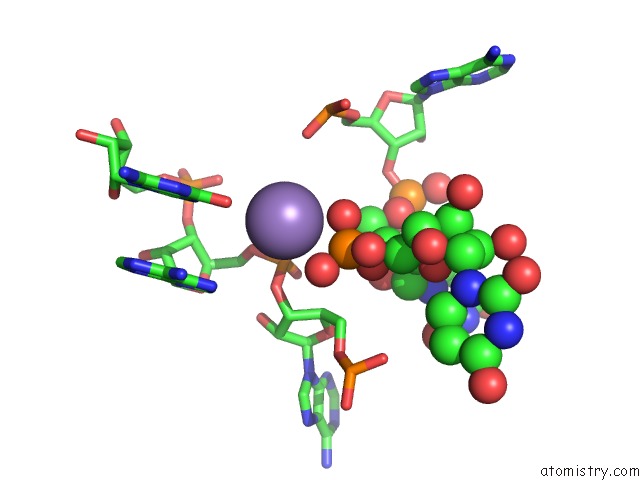
Mono view
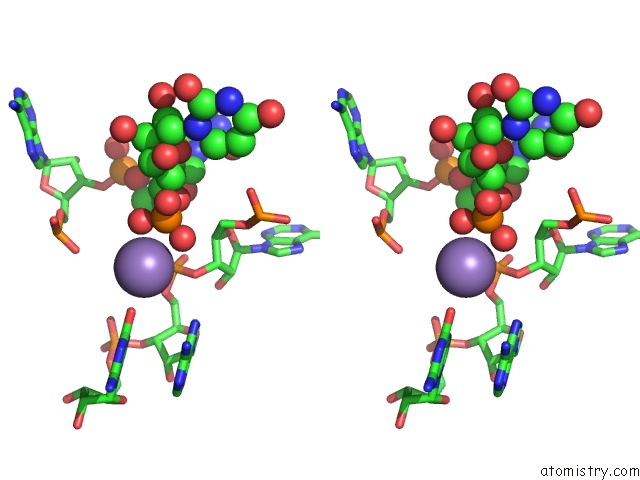
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of Crystal Structure of the T. Tengcongensis Sam-I Riboswitch Variant U34C/A94G Bound with Sam in Manganese Chloride within 5.0Å range:
|
Manganese binding site 5 out of 5 in 3gx6
Go back to
Manganese binding site 5 out
of 5 in the Crystal Structure of the T. Tengcongensis Sam-I Riboswitch Variant U34C/A94G Bound with Sam in Manganese Chloride
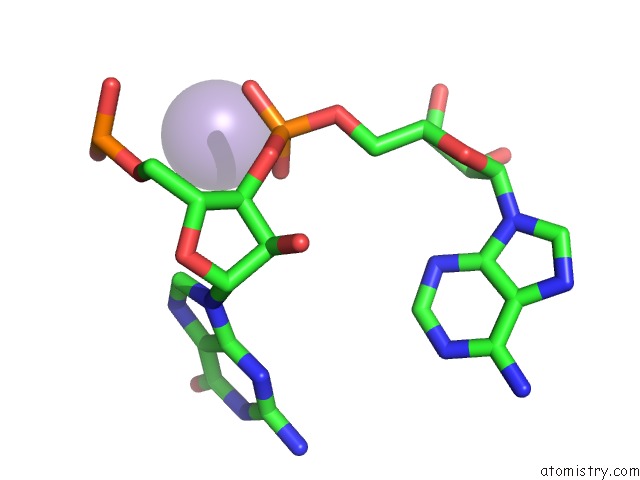
Mono view
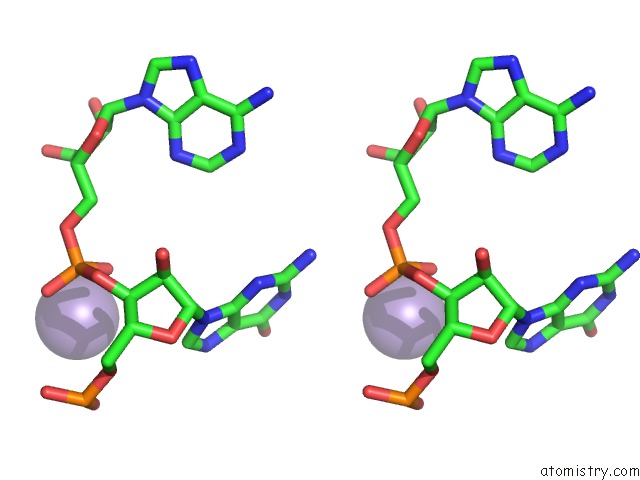
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 5 of Crystal Structure of the T. Tengcongensis Sam-I Riboswitch Variant U34C/A94G Bound with Sam in Manganese Chloride within 5.0Å range:
|
Reference:
R.K.Montange,
E.Mondragon,
D.Van Tyne,
A.D.Garst,
P.Ceres,
R.T.Batey.
Discrimination Between Closely Related Cellular Metabolites By the Sam-I Riboswitch. J.Mol.Biol. V. 396 761 2010.
ISSN: ISSN 0022-2836
PubMed: 20006621
DOI: 10.1016/J.JMB.2009.12.007
Page generated: Sat Aug 16 11:50:45 2025
ISSN: ISSN 0022-2836
PubMed: 20006621
DOI: 10.1016/J.JMB.2009.12.007
Last articles
Mn in 7CJ7Mn in 7CJ6
Mn in 7CJ5
Mn in 7CJ4
Mn in 7C4C
Mn in 7CEC
Mn in 7C4B
Mn in 7C43
Mn in 7C47
Mn in 7BVR