Manganese »
PDB 3cev-3ea3 »
3cu0 »
Manganese in PDB 3cu0: Human Beta 1,3-Glucuronyltransferase I (Glcat-I) in Complex with Udp and Gal-Gal(6-SO4)-Xyl(2-PO4)-O-Ser
Enzymatic activity of Human Beta 1,3-Glucuronyltransferase I (Glcat-I) in Complex with Udp and Gal-Gal(6-SO4)-Xyl(2-PO4)-O-Ser
All present enzymatic activity of Human Beta 1,3-Glucuronyltransferase I (Glcat-I) in Complex with Udp and Gal-Gal(6-SO4)-Xyl(2-PO4)-O-Ser:
2.4.1.135;
2.4.1.135;
Protein crystallography data
The structure of Human Beta 1,3-Glucuronyltransferase I (Glcat-I) in Complex with Udp and Gal-Gal(6-SO4)-Xyl(2-PO4)-O-Ser, PDB code: 3cu0
was solved by
Y.Tone,
L.C.Pedersen,
T.Yamamoto,
H.Kitagawa,
J.Nishihara-Shimizu,
J.Tamura,
M.Negishi,
K.Sugahara,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 23.57 / 1.90 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 57.047, 48.399, 103.708, 90.00, 92.40, 90.00 |
| R / Rfree (%) | 21 / 23.4 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Human Beta 1,3-Glucuronyltransferase I (Glcat-I) in Complex with Udp and Gal-Gal(6-SO4)-Xyl(2-PO4)-O-Ser
(pdb code 3cu0). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 2 binding sites of Manganese where determined in the Human Beta 1,3-Glucuronyltransferase I (Glcat-I) in Complex with Udp and Gal-Gal(6-SO4)-Xyl(2-PO4)-O-Ser, PDB code: 3cu0:
Jump to Manganese binding site number: 1; 2;
In total 2 binding sites of Manganese where determined in the Human Beta 1,3-Glucuronyltransferase I (Glcat-I) in Complex with Udp and Gal-Gal(6-SO4)-Xyl(2-PO4)-O-Ser, PDB code: 3cu0:
Jump to Manganese binding site number: 1; 2;
Manganese binding site 1 out of 2 in 3cu0
Go back to
Manganese binding site 1 out
of 2 in the Human Beta 1,3-Glucuronyltransferase I (Glcat-I) in Complex with Udp and Gal-Gal(6-SO4)-Xyl(2-PO4)-O-Ser
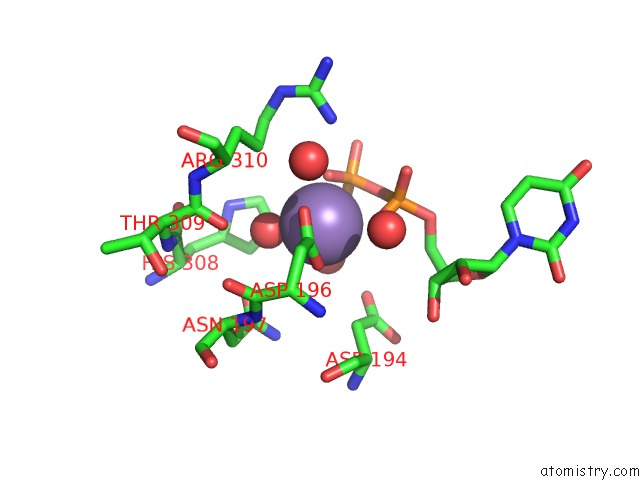
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Human Beta 1,3-Glucuronyltransferase I (Glcat-I) in Complex with Udp and Gal-Gal(6-SO4)-Xyl(2-PO4)-O-Ser within 5.0Å range:
|
Manganese binding site 2 out of 2 in 3cu0
Go back to
Manganese binding site 2 out
of 2 in the Human Beta 1,3-Glucuronyltransferase I (Glcat-I) in Complex with Udp and Gal-Gal(6-SO4)-Xyl(2-PO4)-O-Ser
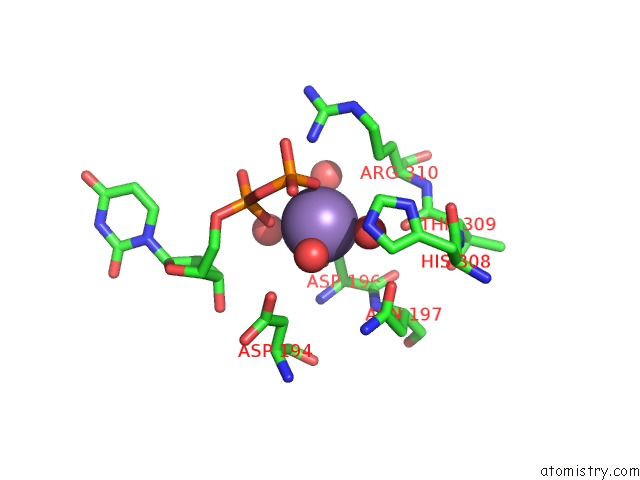
Mono view
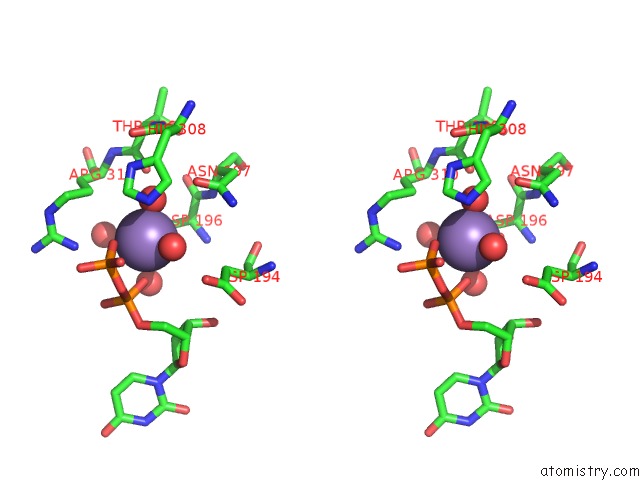
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Human Beta 1,3-Glucuronyltransferase I (Glcat-I) in Complex with Udp and Gal-Gal(6-SO4)-Xyl(2-PO4)-O-Ser within 5.0Å range:
|
Reference:
Y.Tone,
L.C.Pedersen,
T.Yamamoto,
T.Izumikawa,
H.Kitagawa,
J.Nishihara,
J.Tamura,
M.Negishi,
K.Sugahara.
2-O-Phosphorylation of Xylose and 6-O-Sulfation of Galactose in the Protein Linkage Region of Glycosaminoglycans Influence the Glucuronyltransferase-I Activity Involved in the Linkage Region Synthesis. J.Biol.Chem. V. 283 16801 2008.
ISSN: ISSN 0021-9258
PubMed: 18400750
DOI: 10.1074/JBC.M709556200
Page generated: Sat Aug 16 11:34:31 2025
ISSN: ISSN 0021-9258
PubMed: 18400750
DOI: 10.1074/JBC.M709556200
Last articles
Mo in 5O5WMo in 6GB4
Mo in 6GBC
Mo in 6FW2
Mo in 6ETF
Mo in 6GAX
Mo in 6CZA
Mo in 6CZ9
Mo in 6CZ8
Mo in 6CZ7