Manganese »
PDB 3auz-3c5m »
3bg3 »
Manganese in PDB 3bg3: Crystal Structure of Human Pyruvate Carboxylase (Missing the Biotin Carboxylase Domain at the N-Terminus)
Enzymatic activity of Crystal Structure of Human Pyruvate Carboxylase (Missing the Biotin Carboxylase Domain at the N-Terminus)
All present enzymatic activity of Crystal Structure of Human Pyruvate Carboxylase (Missing the Biotin Carboxylase Domain at the N-Terminus):
6.4.1.1;
6.4.1.1;
Protein crystallography data
The structure of Crystal Structure of Human Pyruvate Carboxylase (Missing the Biotin Carboxylase Domain at the N-Terminus), PDB code: 3bg3
was solved by
S.Xiang,
L.Tong,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 2.80 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 81.274, 173.321, 118.194, 90.00, 95.87, 90.00 |
| R / Rfree (%) | 21.6 / 27.1 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Crystal Structure of Human Pyruvate Carboxylase (Missing the Biotin Carboxylase Domain at the N-Terminus)
(pdb code 3bg3). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 4 binding sites of Manganese where determined in the Crystal Structure of Human Pyruvate Carboxylase (Missing the Biotin Carboxylase Domain at the N-Terminus), PDB code: 3bg3:
Jump to Manganese binding site number: 1; 2; 3; 4;
In total 4 binding sites of Manganese where determined in the Crystal Structure of Human Pyruvate Carboxylase (Missing the Biotin Carboxylase Domain at the N-Terminus), PDB code: 3bg3:
Jump to Manganese binding site number: 1; 2; 3; 4;
Manganese binding site 1 out of 4 in 3bg3
Go back to
Manganese binding site 1 out
of 4 in the Crystal Structure of Human Pyruvate Carboxylase (Missing the Biotin Carboxylase Domain at the N-Terminus)
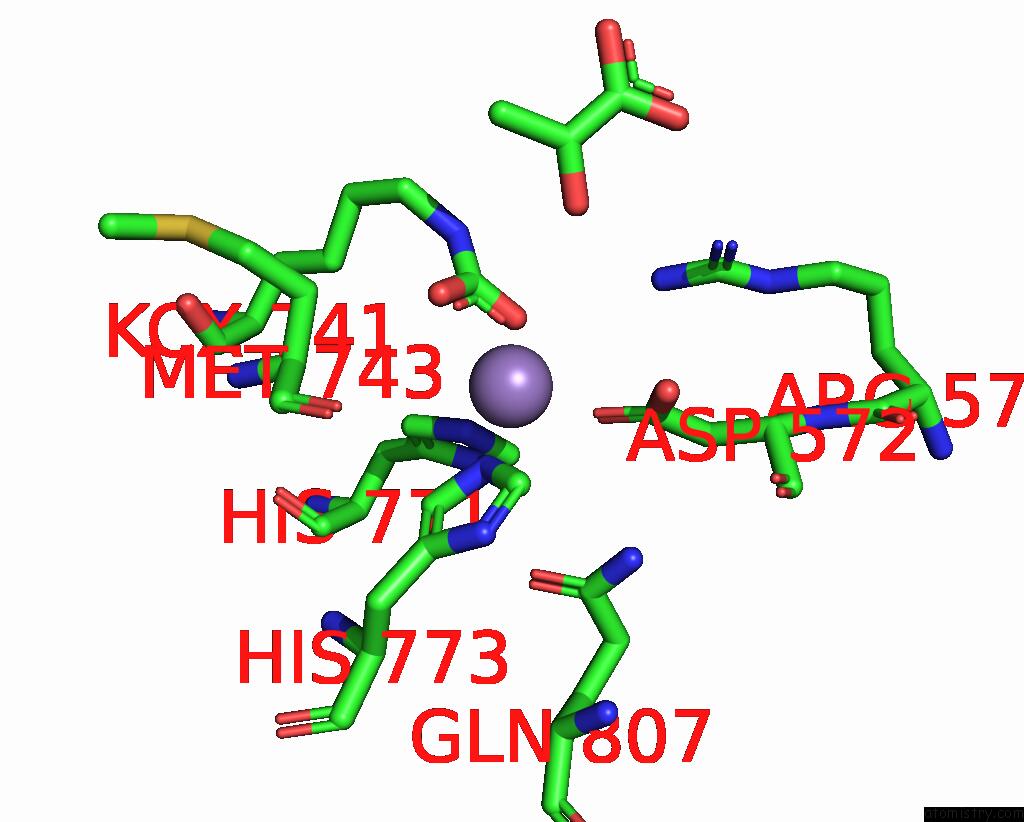
Mono view
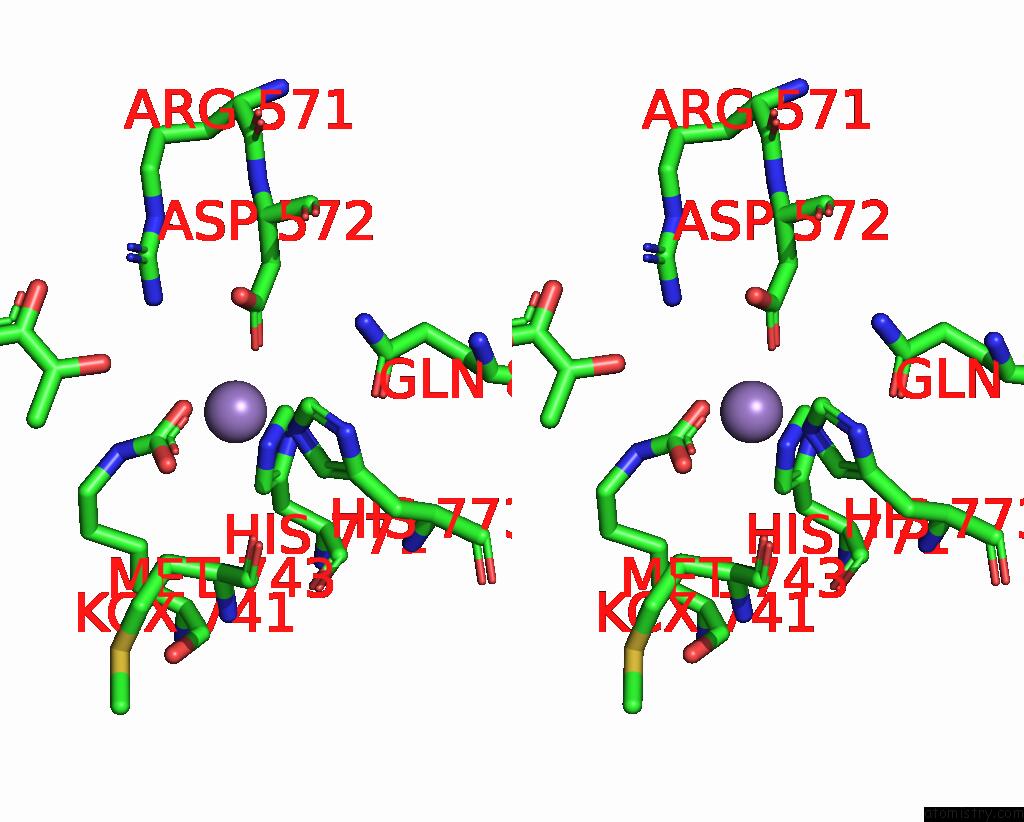
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Crystal Structure of Human Pyruvate Carboxylase (Missing the Biotin Carboxylase Domain at the N-Terminus) within 5.0Å range:
|
Manganese binding site 2 out of 4 in 3bg3
Go back to
Manganese binding site 2 out
of 4 in the Crystal Structure of Human Pyruvate Carboxylase (Missing the Biotin Carboxylase Domain at the N-Terminus)
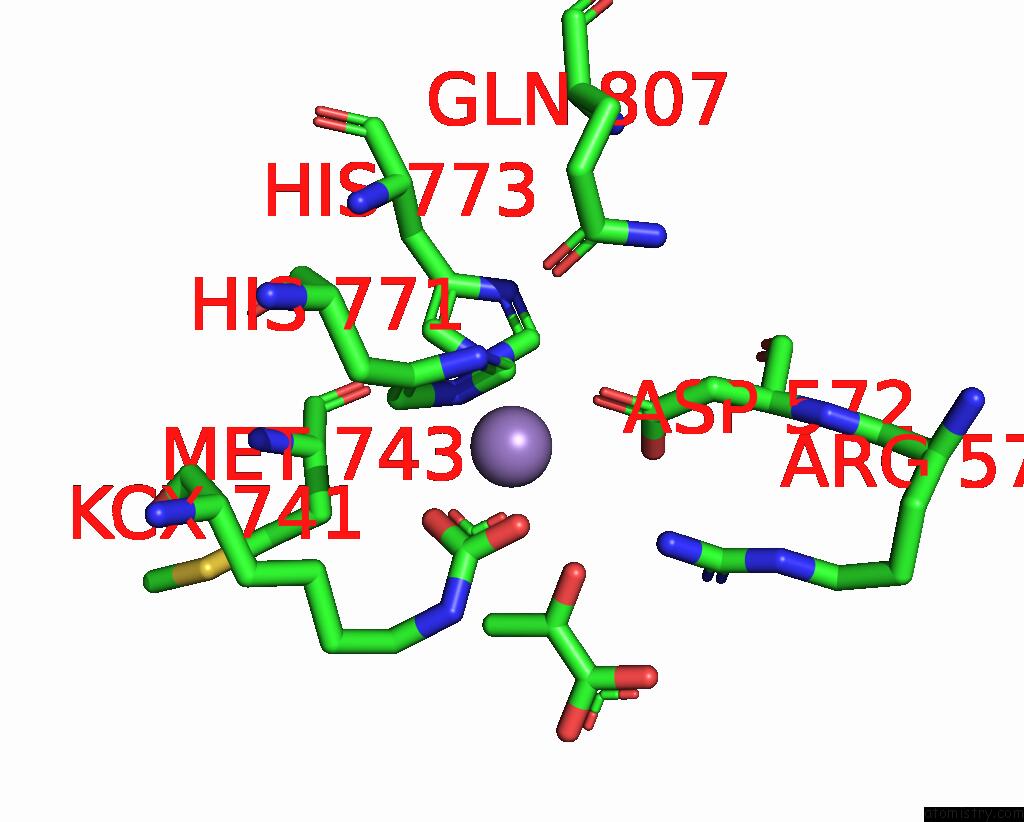
Mono view
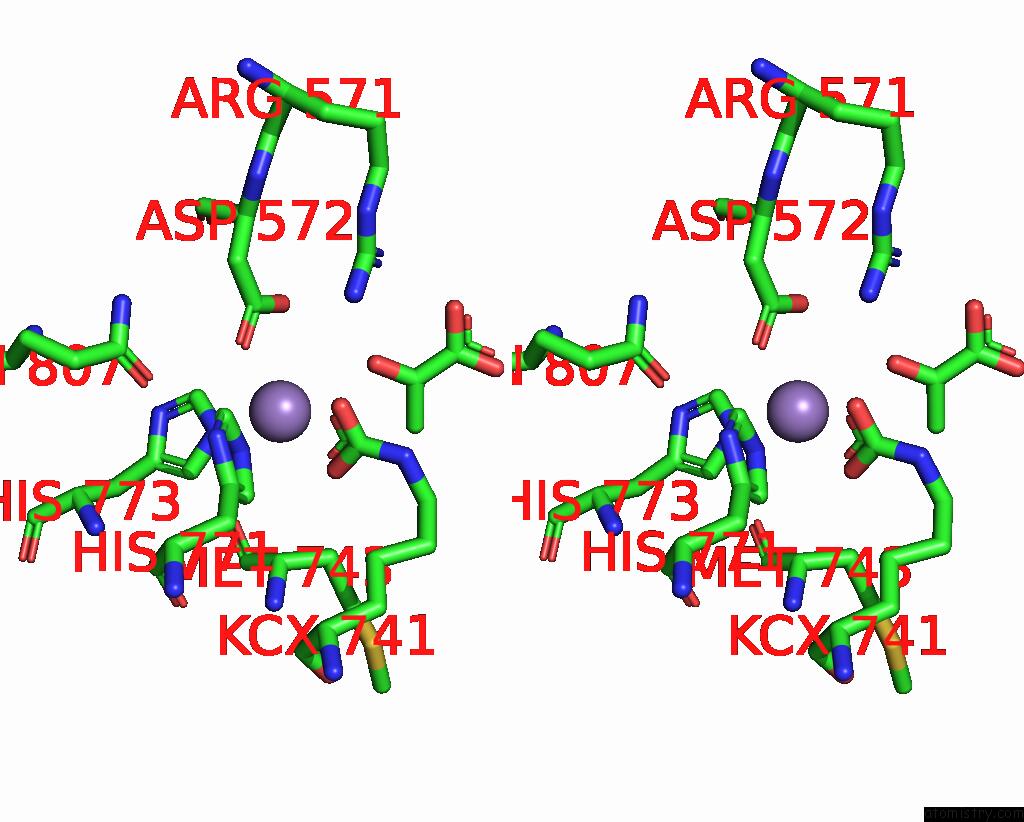
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Crystal Structure of Human Pyruvate Carboxylase (Missing the Biotin Carboxylase Domain at the N-Terminus) within 5.0Å range:
|
Manganese binding site 3 out of 4 in 3bg3
Go back to
Manganese binding site 3 out
of 4 in the Crystal Structure of Human Pyruvate Carboxylase (Missing the Biotin Carboxylase Domain at the N-Terminus)
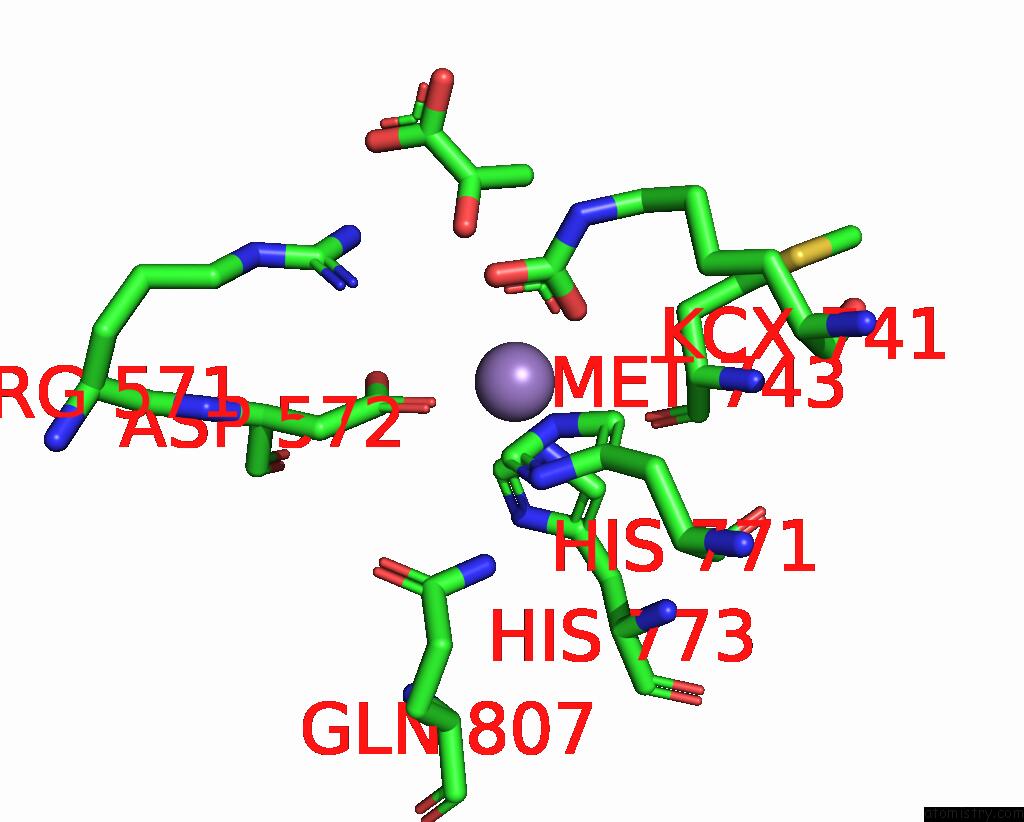
Mono view
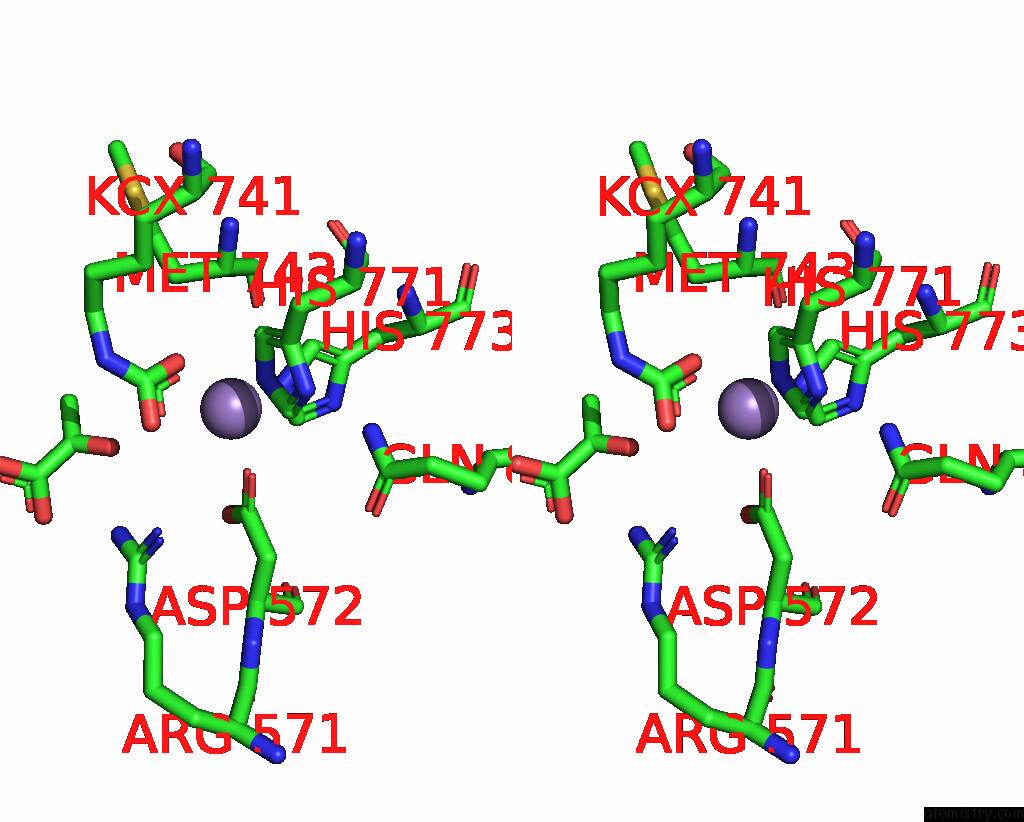
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of Crystal Structure of Human Pyruvate Carboxylase (Missing the Biotin Carboxylase Domain at the N-Terminus) within 5.0Å range:
|
Manganese binding site 4 out of 4 in 3bg3
Go back to
Manganese binding site 4 out
of 4 in the Crystal Structure of Human Pyruvate Carboxylase (Missing the Biotin Carboxylase Domain at the N-Terminus)
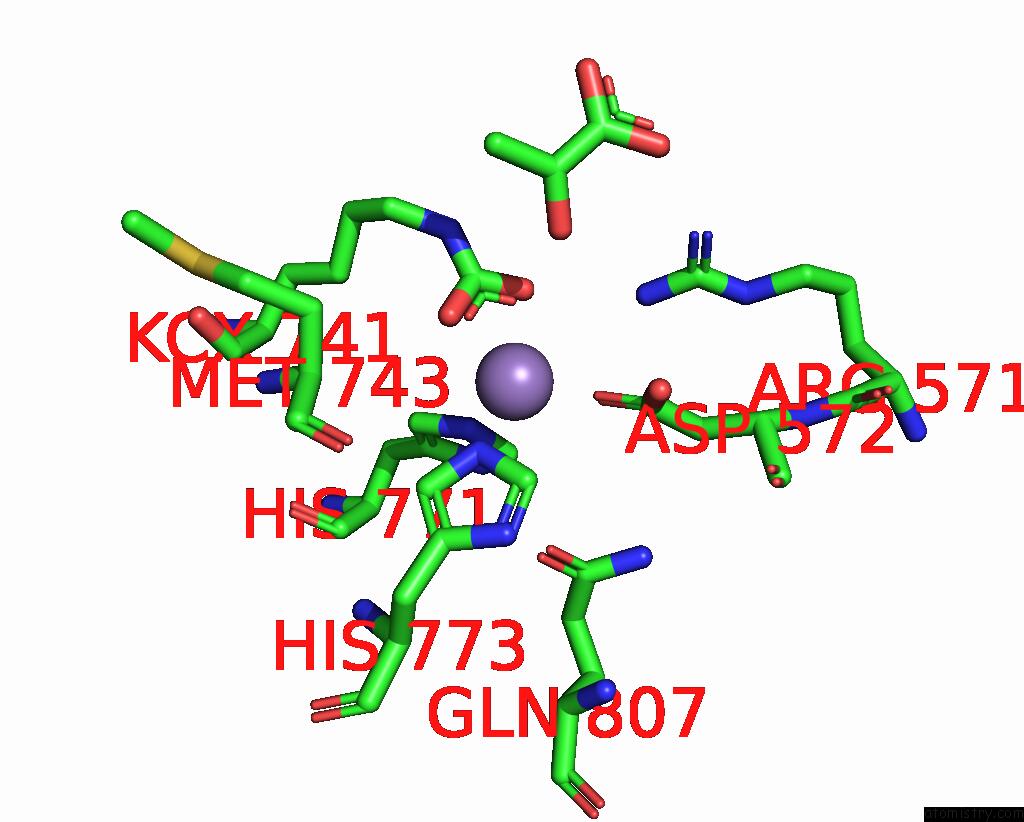
Mono view
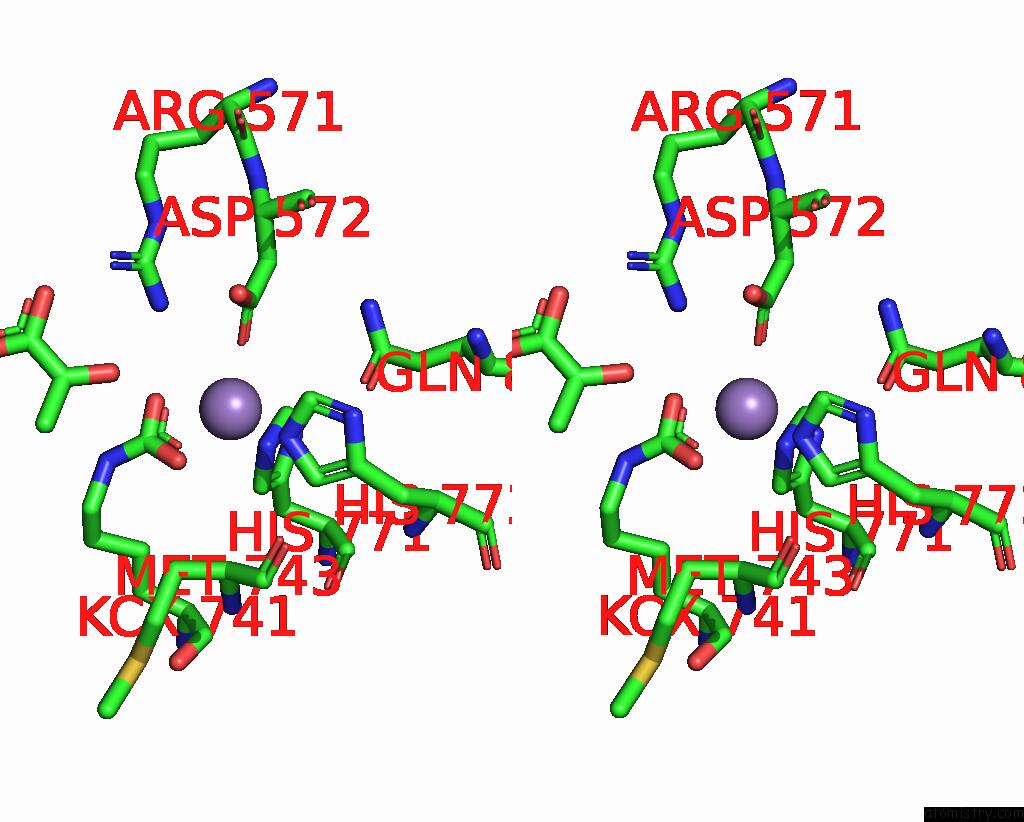
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of Crystal Structure of Human Pyruvate Carboxylase (Missing the Biotin Carboxylase Domain at the N-Terminus) within 5.0Å range:
|
Reference:
S.Xiang,
L.Tong.
Crystal Structures of Human and Staphylococcus Aureus Pyruvate Carboxylase and Molecular Insights Into the Carboxyltransfer Reaction. Nat.Struct.Mol.Biol. V. 15 295 2008.
ISSN: ISSN 1545-9993
PubMed: 18297087
DOI: 10.1038/NSMB.1393
Page generated: Sat Oct 5 15:56:40 2024
ISSN: ISSN 1545-9993
PubMed: 18297087
DOI: 10.1038/NSMB.1393
Last articles
Zn in 9MJ5Zn in 9HNW
Zn in 9G0L
Zn in 9FNE
Zn in 9DZN
Zn in 9E0I
Zn in 9D32
Zn in 9DAK
Zn in 8ZXC
Zn in 8ZUF