Manganese »
PDB 2dvd-2fer »
2fer »
Manganese in PDB 2fer: P450CAM From Pseudomonas Putida Reconstituted with Manganic Protoporphyrin IX
Enzymatic activity of P450CAM From Pseudomonas Putida Reconstituted with Manganic Protoporphyrin IX
All present enzymatic activity of P450CAM From Pseudomonas Putida Reconstituted with Manganic Protoporphyrin IX:
1.14.15.1;
1.14.15.1;
Protein crystallography data
The structure of P450CAM From Pseudomonas Putida Reconstituted with Manganic Protoporphyrin IX, PDB code: 2fer
was solved by
K.Von Koenig,
T.M.Makris,
S.G.Sligar,
I.Schlichting,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 17.59 / 1.70 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 63.320, 63.820, 105.440, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.9 / 22.5 |
Other elements in 2fer:
The structure of P450CAM From Pseudomonas Putida Reconstituted with Manganic Protoporphyrin IX also contains other interesting chemical elements:
| Potassium | (K) | 1 atom |
Manganese Binding Sites:
The binding sites of Manganese atom in the P450CAM From Pseudomonas Putida Reconstituted with Manganic Protoporphyrin IX
(pdb code 2fer). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total only one binding site of Manganese was determined in the P450CAM From Pseudomonas Putida Reconstituted with Manganic Protoporphyrin IX, PDB code: 2fer:
In total only one binding site of Manganese was determined in the P450CAM From Pseudomonas Putida Reconstituted with Manganic Protoporphyrin IX, PDB code: 2fer:
Manganese binding site 1 out of 1 in 2fer
Go back to
Manganese binding site 1 out
of 1 in the P450CAM From Pseudomonas Putida Reconstituted with Manganic Protoporphyrin IX
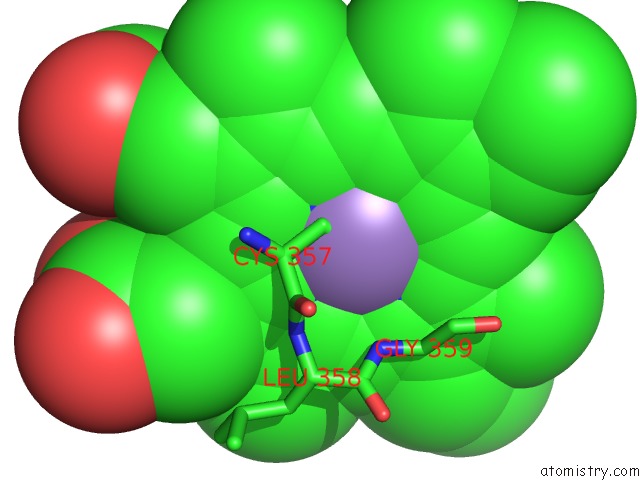
Mono view
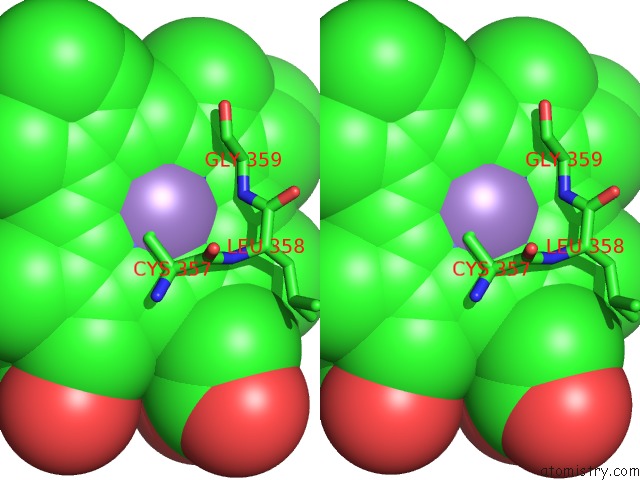
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of P450CAM From Pseudomonas Putida Reconstituted with Manganic Protoporphyrin IX within 5.0Å range:
|
Reference:
T.M.Makris,
K.Von Koenig,
I.Schlichting,
S.G.Sligar.
The Status of High-Valent Metal Oxo Complexes in the P450 Cytochromes. J.Inorg.Biochem. V. 100 507 2006.
ISSN: ISSN 0162-0134
PubMed: 16510191
DOI: 10.1016/J.JINORGBIO.2006.01.025
Page generated: Sat Aug 16 10:12:02 2025
ISSN: ISSN 0162-0134
PubMed: 16510191
DOI: 10.1016/J.JINORGBIO.2006.01.025
Last articles
Na in 1NXDNa in 1NVJ
Na in 1NZA
Na in 1NTB
Na in 1NTA
Na in 1NSZ
Na in 1NSX
Na in 1NSV
Na in 1NSU
Na in 1NSS