Manganese »
PDB 2dvd-2fer »
2f6k »
Manganese in PDB 2f6k: Crystal Structure of Amidohydrorolase II; Northeast Structural Genomics Target LPR24
Enzymatic activity of Crystal Structure of Amidohydrorolase II; Northeast Structural Genomics Target LPR24
All present enzymatic activity of Crystal Structure of Amidohydrorolase II; Northeast Structural Genomics Target LPR24:
4.1.1.45;
4.1.1.45;
Protein crystallography data
The structure of Crystal Structure of Amidohydrorolase II; Northeast Structural Genomics Target LPR24, PDB code: 2f6k
was solved by
K.Das,
R.Xiao,
T.Acton,
L.Ma,
E.Arnold,
G.T.Montelione,
Northeaststructural Genomics Consortium (Nesg),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 49.90 / 2.50 |
| Space group | P 43 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 82.620, 82.620, 240.815, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.2 / 22.5 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Crystal Structure of Amidohydrorolase II; Northeast Structural Genomics Target LPR24
(pdb code 2f6k). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 2 binding sites of Manganese where determined in the Crystal Structure of Amidohydrorolase II; Northeast Structural Genomics Target LPR24, PDB code: 2f6k:
Jump to Manganese binding site number: 1; 2;
In total 2 binding sites of Manganese where determined in the Crystal Structure of Amidohydrorolase II; Northeast Structural Genomics Target LPR24, PDB code: 2f6k:
Jump to Manganese binding site number: 1; 2;
Manganese binding site 1 out of 2 in 2f6k
Go back to
Manganese binding site 1 out
of 2 in the Crystal Structure of Amidohydrorolase II; Northeast Structural Genomics Target LPR24
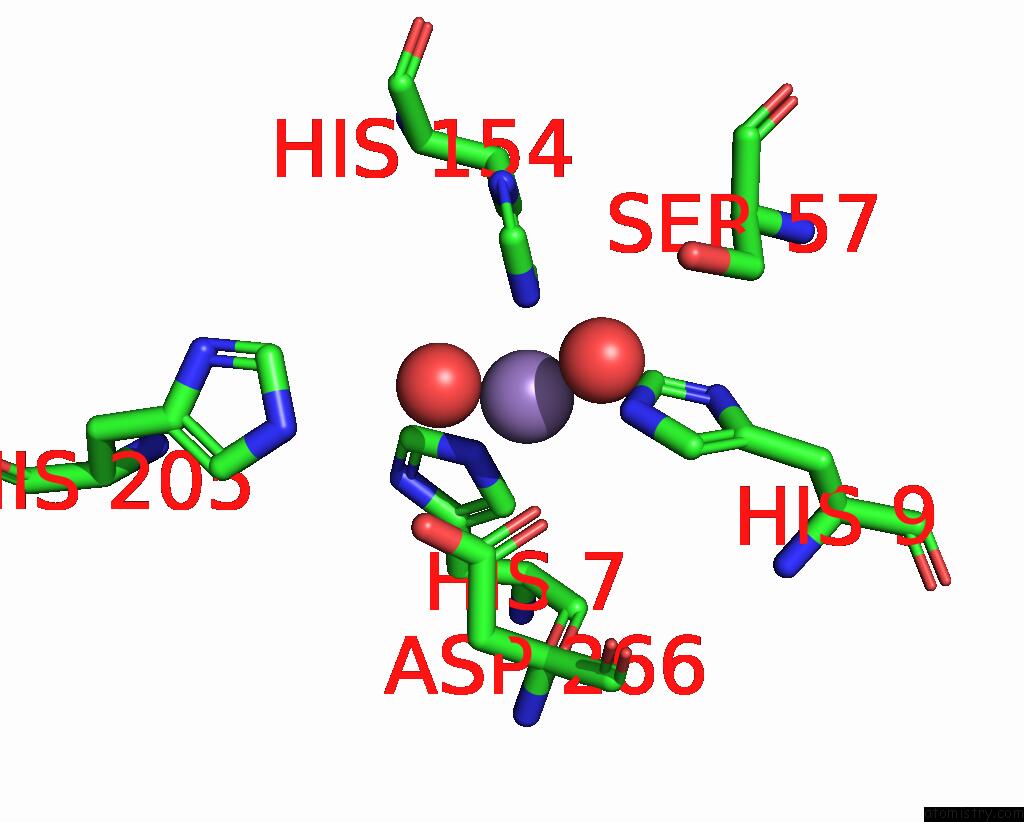
Mono view
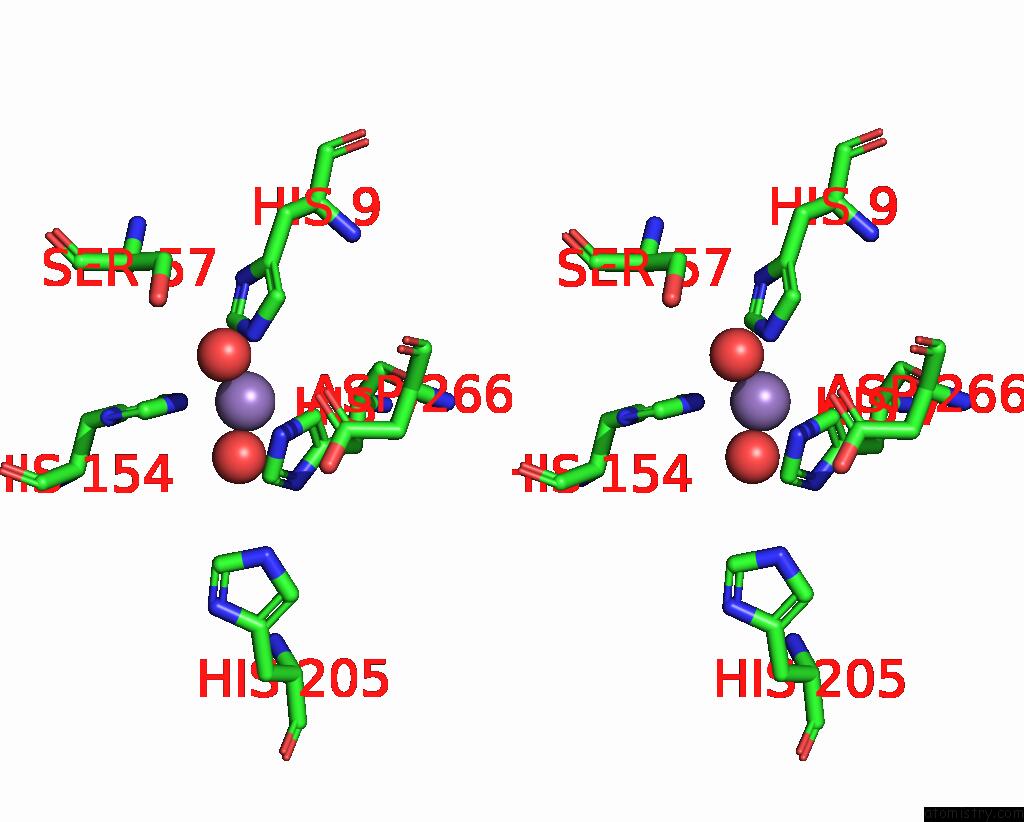
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Crystal Structure of Amidohydrorolase II; Northeast Structural Genomics Target LPR24 within 5.0Å range:
|
Manganese binding site 2 out of 2 in 2f6k
Go back to
Manganese binding site 2 out
of 2 in the Crystal Structure of Amidohydrorolase II; Northeast Structural Genomics Target LPR24
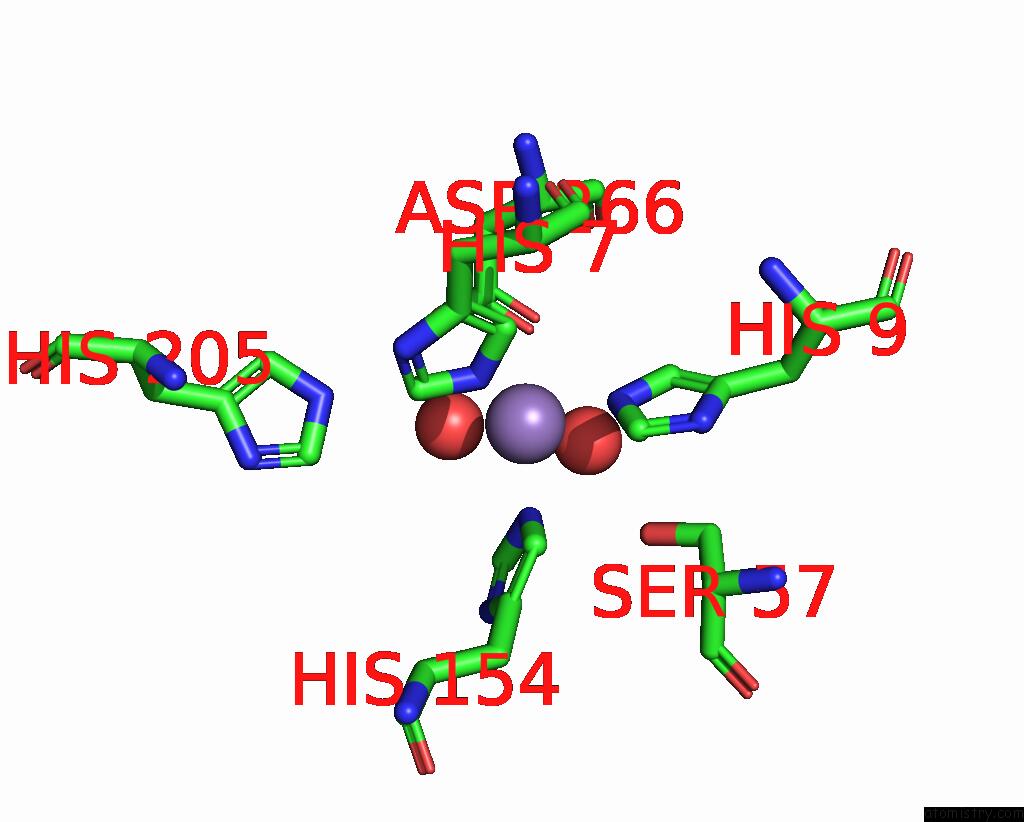
Mono view
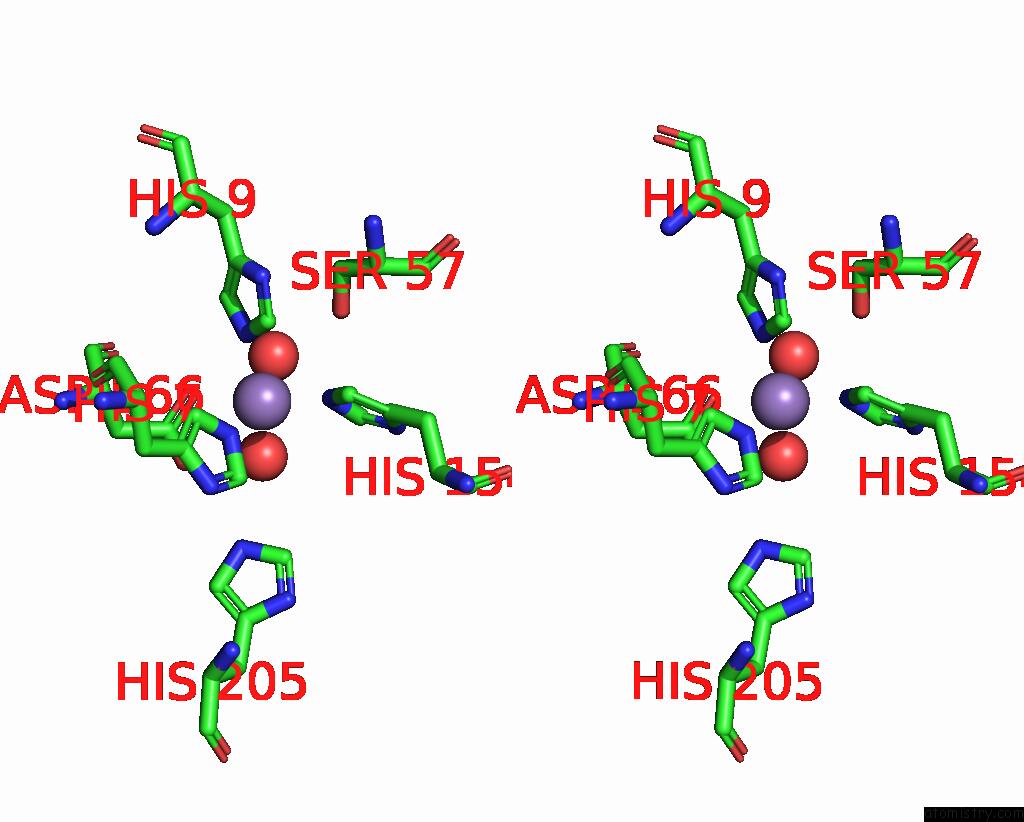
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Crystal Structure of Amidohydrorolase II; Northeast Structural Genomics Target LPR24 within 5.0Å range:
|
Reference:
K.Das,
R.Xiao,
T.Acton,
L.Ma,
E.Arnold,
G.T.Montelione.
Crystal Structure of Acmds From Lactobacillus Plantarum To Be Published.
Page generated: Sat Oct 5 14:02:15 2024
Last articles
F in 7LAYF in 7LAJ
F in 7LAE
F in 7L9M
F in 7L9J
F in 7LAD
F in 7L8J
F in 7L8I
F in 7L7N
F in 7L8H