Manganese »
PDB 2a7a-2axt »
2a8r »
Manganese in PDB 2a8r: 2.45 Angstrom Crystal Structure of the Complex Between the Nuclear Snorna Decapping Nudix Hydrolase X29 and Manganese in the Presence of 7-Methyl-Gtp
Protein crystallography data
The structure of 2.45 Angstrom Crystal Structure of the Complex Between the Nuclear Snorna Decapping Nudix Hydrolase X29 and Manganese in the Presence of 7-Methyl-Gtp, PDB code: 2a8r
was solved by
J.N.Scarsdale,
B.A.Peculis,
H.T.Wright,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 23.00 / 2.45 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 50.200, 82.150, 112.240, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 22.2 / 26.3 |
Manganese Binding Sites:
The binding sites of Manganese atom in the 2.45 Angstrom Crystal Structure of the Complex Between the Nuclear Snorna Decapping Nudix Hydrolase X29 and Manganese in the Presence of 7-Methyl-Gtp
(pdb code 2a8r). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 5 binding sites of Manganese where determined in the 2.45 Angstrom Crystal Structure of the Complex Between the Nuclear Snorna Decapping Nudix Hydrolase X29 and Manganese in the Presence of 7-Methyl-Gtp, PDB code: 2a8r:
Jump to Manganese binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Manganese where determined in the 2.45 Angstrom Crystal Structure of the Complex Between the Nuclear Snorna Decapping Nudix Hydrolase X29 and Manganese in the Presence of 7-Methyl-Gtp, PDB code: 2a8r:
Jump to Manganese binding site number: 1; 2; 3; 4; 5;
Manganese binding site 1 out of 5 in 2a8r
Go back to
Manganese binding site 1 out
of 5 in the 2.45 Angstrom Crystal Structure of the Complex Between the Nuclear Snorna Decapping Nudix Hydrolase X29 and Manganese in the Presence of 7-Methyl-Gtp
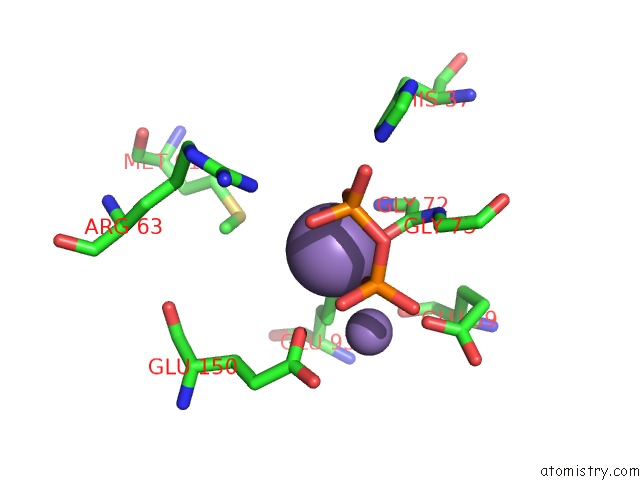
Mono view
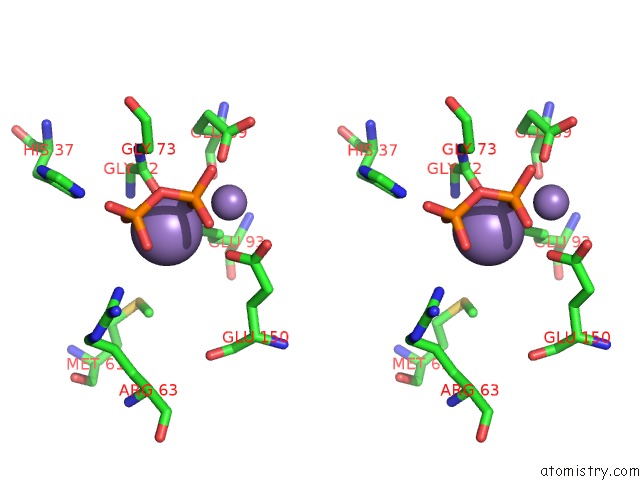
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of 2.45 Angstrom Crystal Structure of the Complex Between the Nuclear Snorna Decapping Nudix Hydrolase X29 and Manganese in the Presence of 7-Methyl-Gtp within 5.0Å range:
|
Manganese binding site 2 out of 5 in 2a8r
Go back to
Manganese binding site 2 out
of 5 in the 2.45 Angstrom Crystal Structure of the Complex Between the Nuclear Snorna Decapping Nudix Hydrolase X29 and Manganese in the Presence of 7-Methyl-Gtp
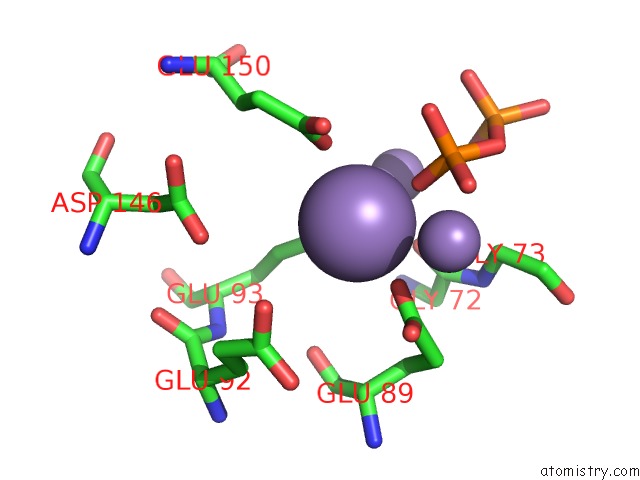
Mono view
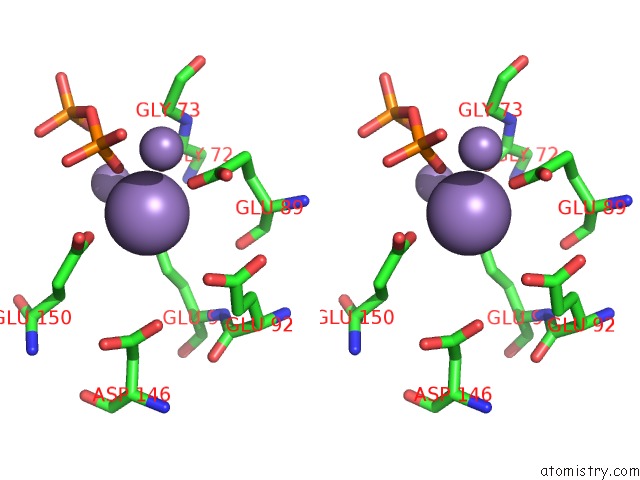
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of 2.45 Angstrom Crystal Structure of the Complex Between the Nuclear Snorna Decapping Nudix Hydrolase X29 and Manganese in the Presence of 7-Methyl-Gtp within 5.0Å range:
|
Manganese binding site 3 out of 5 in 2a8r
Go back to
Manganese binding site 3 out
of 5 in the 2.45 Angstrom Crystal Structure of the Complex Between the Nuclear Snorna Decapping Nudix Hydrolase X29 and Manganese in the Presence of 7-Methyl-Gtp
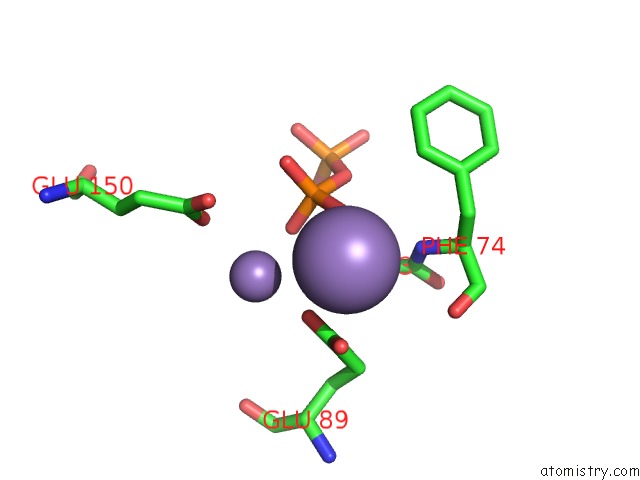
Mono view
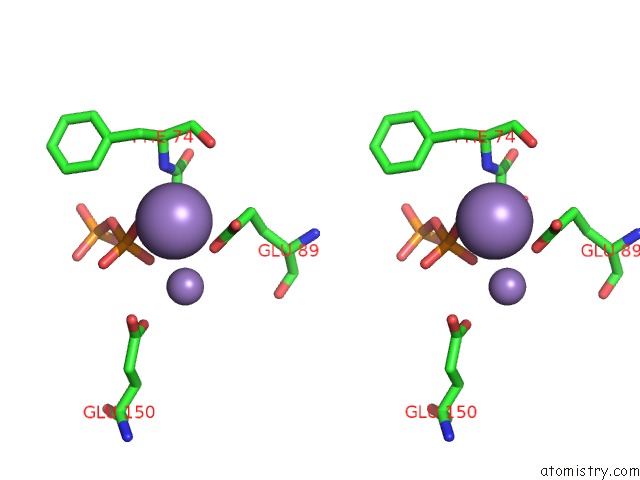
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 3 of 2.45 Angstrom Crystal Structure of the Complex Between the Nuclear Snorna Decapping Nudix Hydrolase X29 and Manganese in the Presence of 7-Methyl-Gtp within 5.0Å range:
|
Manganese binding site 4 out of 5 in 2a8r
Go back to
Manganese binding site 4 out
of 5 in the 2.45 Angstrom Crystal Structure of the Complex Between the Nuclear Snorna Decapping Nudix Hydrolase X29 and Manganese in the Presence of 7-Methyl-Gtp
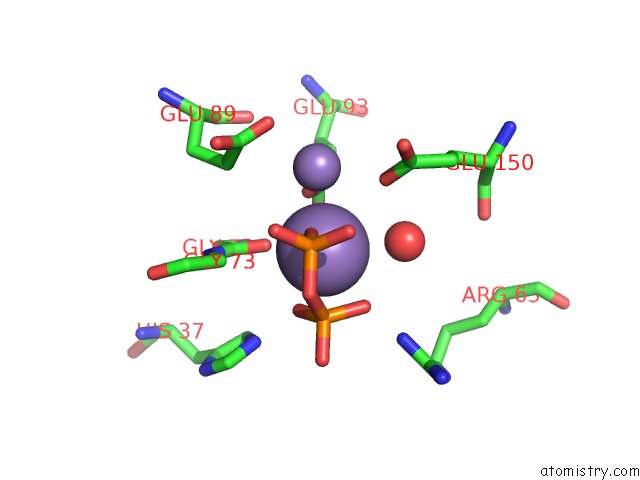
Mono view
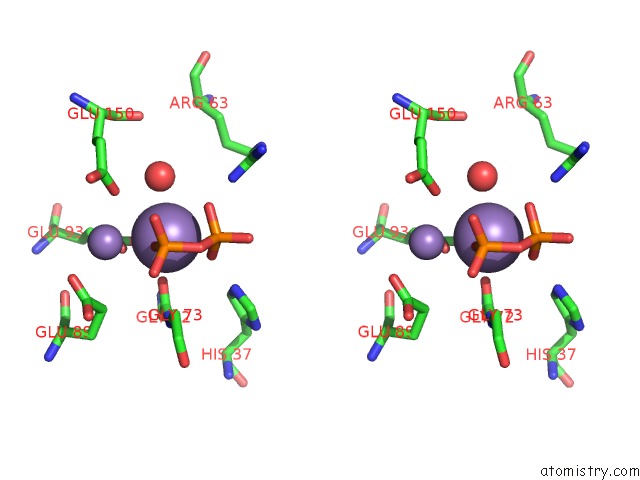
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 4 of 2.45 Angstrom Crystal Structure of the Complex Between the Nuclear Snorna Decapping Nudix Hydrolase X29 and Manganese in the Presence of 7-Methyl-Gtp within 5.0Å range:
|
Manganese binding site 5 out of 5 in 2a8r
Go back to
Manganese binding site 5 out
of 5 in the 2.45 Angstrom Crystal Structure of the Complex Between the Nuclear Snorna Decapping Nudix Hydrolase X29 and Manganese in the Presence of 7-Methyl-Gtp
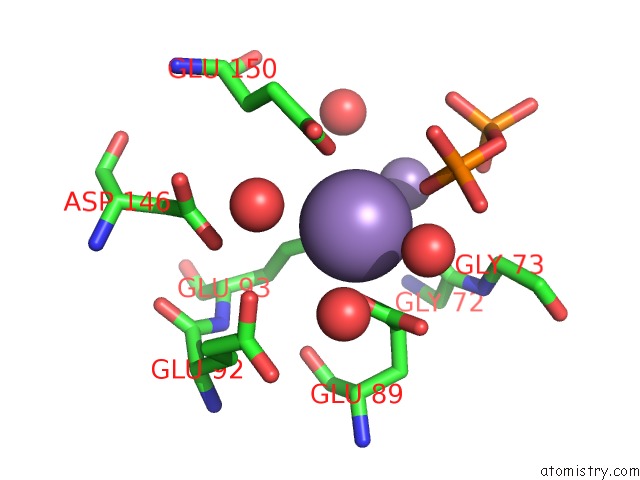
Mono view
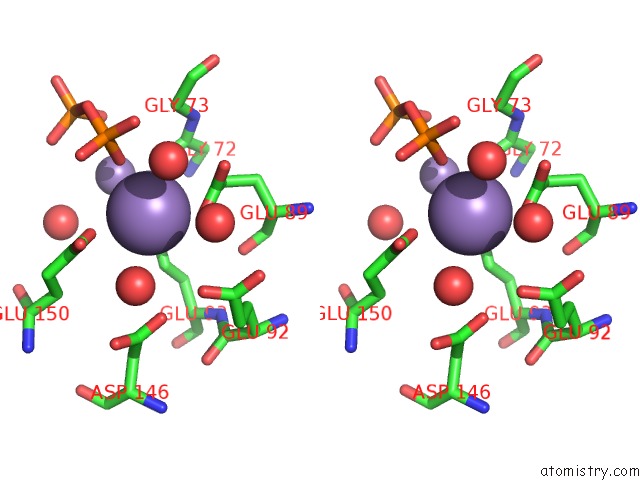
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 5 of 2.45 Angstrom Crystal Structure of the Complex Between the Nuclear Snorna Decapping Nudix Hydrolase X29 and Manganese in the Presence of 7-Methyl-Gtp within 5.0Å range:
|
Reference:
J.N.Scarsdale,
B.A.Peculis,
H.T.Wright.
Crystal Structures of U8 Snorna Decapping Nudix Hydrolase, X29, and Its Metal and Cap Complexes Structure V. 14 331 2006.
ISSN: ISSN 0969-2126
PubMed: 16472752
DOI: 10.1016/J.STR.2005.11.010
Page generated: Sat Aug 16 09:40:42 2025
ISSN: ISSN 0969-2126
PubMed: 16472752
DOI: 10.1016/J.STR.2005.11.010
Last articles
Na in 1JZ2Na in 1JYX
Na in 1JYW
Na in 1JYV
Na in 1JYN
Na in 1JTW
Na in 1JTT
Na in 1JTP
Na in 1JRZ
Na in 1JRY