Manganese »
PDB 1n0n-1o99 »
1nr0 »
Manganese in PDB 1nr0: Two Seven-Bladed Beta-Propeller Domains Revealed By the Structure of A C. Elegans Homologue of Yeast Actin Interacting Protein 1 (AIP1).
Protein crystallography data
The structure of Two Seven-Bladed Beta-Propeller Domains Revealed By the Structure of A C. Elegans Homologue of Yeast Actin Interacting Protein 1 (AIP1)., PDB code: 1nr0
was solved by
S.M.Vorobiev,
K.Mohri,
S.Ono,
S.C.Almo,
S.K.Burley,
New York Sgxresearch Center For Structural Genomics (Nysgxrc),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 23.59 / 1.70 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 64.845, 65.174, 69.240, 90.00, 98.28, 90.00 |
| R / Rfree (%) | 19.9 / 23 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Two Seven-Bladed Beta-Propeller Domains Revealed By the Structure of A C. Elegans Homologue of Yeast Actin Interacting Protein 1 (AIP1).
(pdb code 1nr0). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total only one binding site of Manganese was determined in the Two Seven-Bladed Beta-Propeller Domains Revealed By the Structure of A C. Elegans Homologue of Yeast Actin Interacting Protein 1 (AIP1)., PDB code: 1nr0:
In total only one binding site of Manganese was determined in the Two Seven-Bladed Beta-Propeller Domains Revealed By the Structure of A C. Elegans Homologue of Yeast Actin Interacting Protein 1 (AIP1)., PDB code: 1nr0:
Manganese binding site 1 out of 1 in 1nr0
Go back to
Manganese binding site 1 out
of 1 in the Two Seven-Bladed Beta-Propeller Domains Revealed By the Structure of A C. Elegans Homologue of Yeast Actin Interacting Protein 1 (AIP1).
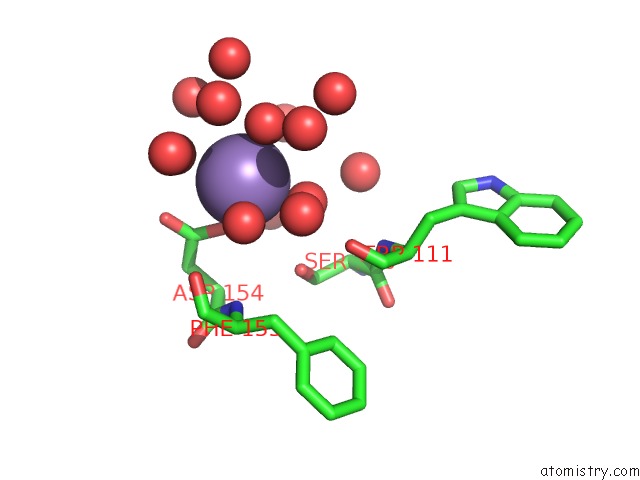
Mono view
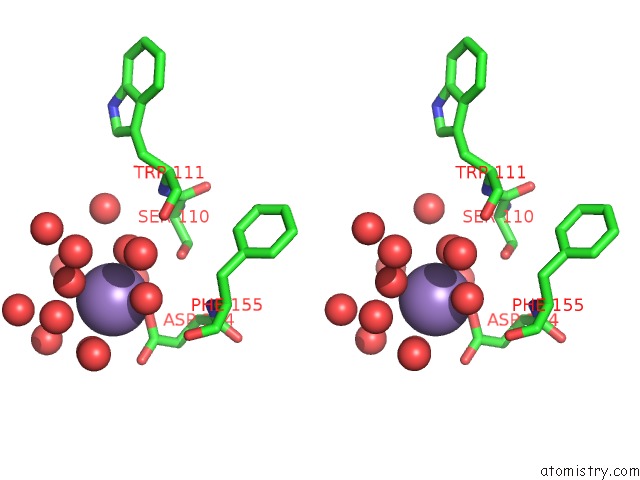
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Two Seven-Bladed Beta-Propeller Domains Revealed By the Structure of A C. Elegans Homologue of Yeast Actin Interacting Protein 1 (AIP1). within 5.0Å range:
|
Reference:
K.Mohri,
S.M.Vorobiev,
A.A.Fedorov,
S.C.Almo,
S.Ono.
Identification of Functional Residues on Caenorhabditis Elegans Actin-Interacting Protein 1 (Unc-78) For Disassembly of Actin Depolymerizing Factor/Cofilin-Bound Actin Filaments J.Biol.Chem. V. 279 31697 2004.
ISSN: ISSN 0021-9258
PubMed: 15150269
DOI: 10.1074/JBC.M403351200
Page generated: Sat Oct 5 11:53:08 2024
ISSN: ISSN 0021-9258
PubMed: 15150269
DOI: 10.1074/JBC.M403351200
Last articles
Ca in 5MNKCa in 5MNH
Ca in 5MNG
Ca in 5MNF
Ca in 5MNE
Ca in 5MNC
Ca in 5MNB
Ca in 5MKG
Ca in 5MN1
Ca in 5MNA