Manganese »
PDB 1j54-1khb »
1jn2 »
Manganese in PDB 1jn2: Crystal Structure of Meso-Tetrasulphonatophenyl Porphyrin Complexed with Concanavalin A
Protein crystallography data
The structure of Crystal Structure of Meso-Tetrasulphonatophenyl Porphyrin Complexed with Concanavalin A, PDB code: 1jn2
was solved by
M.Goel,
D.Jain,
K.J.Kaur,
R.Kenoth,
B.G.Maiya,
M.J.Swamy,
D.M.Salunke,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 100.00 / 1.90 |
| Space group | F 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 106.000, 117.300, 126.000, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.5 / 23.2 |
Other elements in 1jn2:
The structure of Crystal Structure of Meso-Tetrasulphonatophenyl Porphyrin Complexed with Concanavalin A also contains other interesting chemical elements:
| Calcium | (Ca) | 1 atom |
Manganese Binding Sites:
The binding sites of Manganese atom in the Crystal Structure of Meso-Tetrasulphonatophenyl Porphyrin Complexed with Concanavalin A
(pdb code 1jn2). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total only one binding site of Manganese was determined in the Crystal Structure of Meso-Tetrasulphonatophenyl Porphyrin Complexed with Concanavalin A, PDB code: 1jn2:
In total only one binding site of Manganese was determined in the Crystal Structure of Meso-Tetrasulphonatophenyl Porphyrin Complexed with Concanavalin A, PDB code: 1jn2:
Manganese binding site 1 out of 1 in 1jn2
Go back to
Manganese binding site 1 out
of 1 in the Crystal Structure of Meso-Tetrasulphonatophenyl Porphyrin Complexed with Concanavalin A
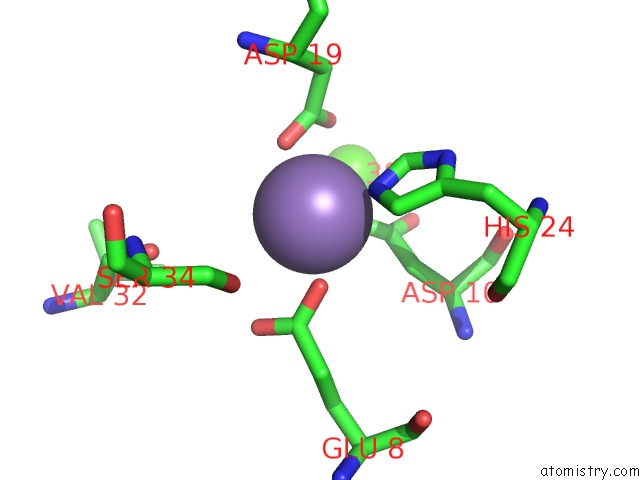
Mono view
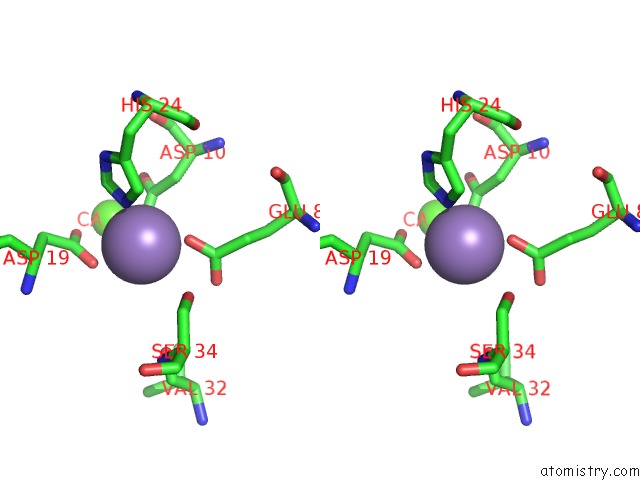
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Crystal Structure of Meso-Tetrasulphonatophenyl Porphyrin Complexed with Concanavalin A within 5.0Å range:
|
Reference:
M.Goel,
D.Jain,
K.J.Kaur,
R.Kenoth,
B.G.Maiya,
M.J.Swamy,
D.M.Salunke.
Functional Equality in the Absence of Structural Similarity: An Added Dimension to Molecular Mimicry J.Biol.Chem. V. 276 39277 2000.
ISSN: ISSN 0021-9258
PubMed: 11504727
DOI: 10.1074/JBC.M105387200
Page generated: Sat Oct 5 11:12:25 2024
ISSN: ISSN 0021-9258
PubMed: 11504727
DOI: 10.1074/JBC.M105387200
Last articles
Cl in 7TSNCl in 7TUO
Cl in 7TSM
Cl in 7TSL
Cl in 7TSK
Cl in 7TRV
Cl in 7TSI
Cl in 7TSG
Cl in 7TSH
Cl in 7TRU