Manganese »
PDB 1ho5-1j53 »
1j53 »
Manganese in PDB 1j53: Structure of the N-Terminal Exonuclease Domain of the Epsilon Subunit of E.Coli Dna Polymerase III at pH 8.5
Enzymatic activity of Structure of the N-Terminal Exonuclease Domain of the Epsilon Subunit of E.Coli Dna Polymerase III at pH 8.5
All present enzymatic activity of Structure of the N-Terminal Exonuclease Domain of the Epsilon Subunit of E.Coli Dna Polymerase III at pH 8.5:
2.7.7.7;
2.7.7.7;
Protein crystallography data
The structure of Structure of the N-Terminal Exonuclease Domain of the Epsilon Subunit of E.Coli Dna Polymerase III at pH 8.5, PDB code: 1j53
was solved by
S.Hamdan,
P.D.Carr,
S.E.Brown,
D.L.Ollis,
N.E.Dixon,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 1.80 |
| Space group | P 41 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 60.800, 60.800, 111.100, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.9 / 22.7 |
Manganese Binding Sites:
The binding sites of Manganese atom in the Structure of the N-Terminal Exonuclease Domain of the Epsilon Subunit of E.Coli Dna Polymerase III at pH 8.5
(pdb code 1j53). This binding sites where shown within
5.0 Angstroms radius around Manganese atom.
In total 2 binding sites of Manganese where determined in the Structure of the N-Terminal Exonuclease Domain of the Epsilon Subunit of E.Coli Dna Polymerase III at pH 8.5, PDB code: 1j53:
Jump to Manganese binding site number: 1; 2;
In total 2 binding sites of Manganese where determined in the Structure of the N-Terminal Exonuclease Domain of the Epsilon Subunit of E.Coli Dna Polymerase III at pH 8.5, PDB code: 1j53:
Jump to Manganese binding site number: 1; 2;
Manganese binding site 1 out of 2 in 1j53
Go back to
Manganese binding site 1 out
of 2 in the Structure of the N-Terminal Exonuclease Domain of the Epsilon Subunit of E.Coli Dna Polymerase III at pH 8.5
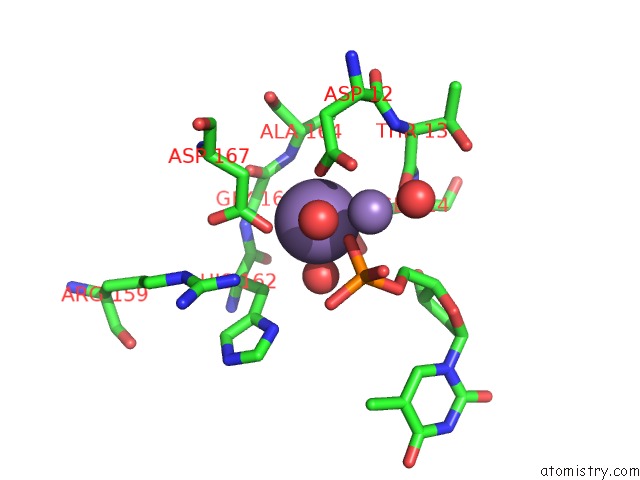
Mono view
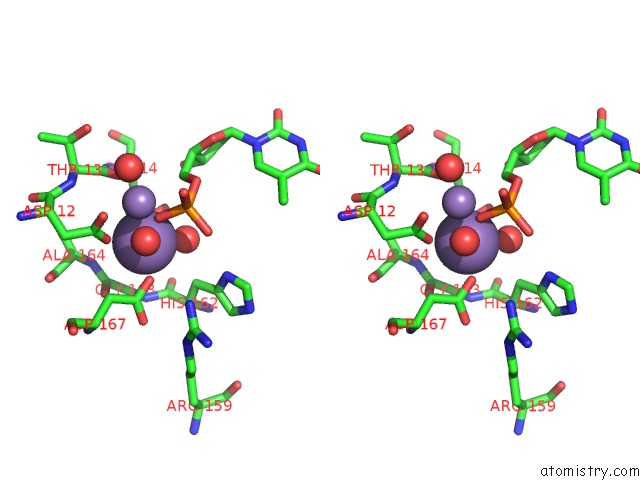
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 1 of Structure of the N-Terminal Exonuclease Domain of the Epsilon Subunit of E.Coli Dna Polymerase III at pH 8.5 within 5.0Å range:
|
Manganese binding site 2 out of 2 in 1j53
Go back to
Manganese binding site 2 out
of 2 in the Structure of the N-Terminal Exonuclease Domain of the Epsilon Subunit of E.Coli Dna Polymerase III at pH 8.5
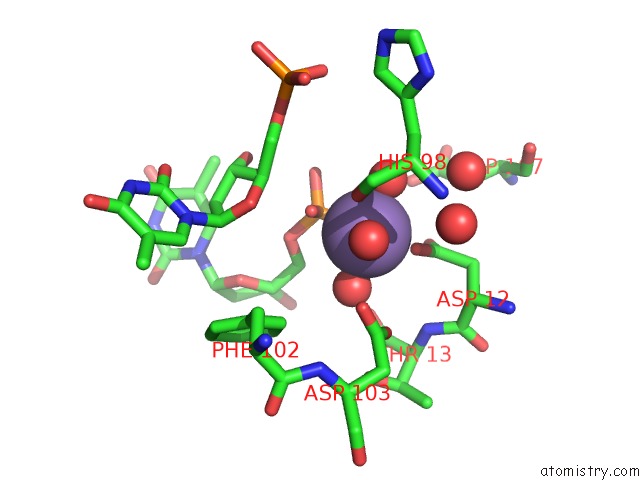
Mono view
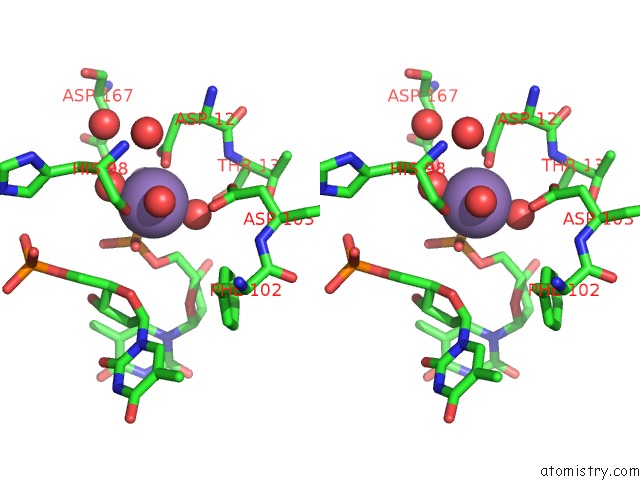
Stereo pair view

Mono view

Stereo pair view
A full contact list of Manganese with other atoms in the Mn binding
site number 2 of Structure of the N-Terminal Exonuclease Domain of the Epsilon Subunit of E.Coli Dna Polymerase III at pH 8.5 within 5.0Å range:
|
Reference:
S.Hamdan,
P.D.Carr,
S.E.Brown,
D.L.Ollis,
N.E.Dixon.
Structural Basis For Proofreading During Replication of the Escherichia Coli Chromosome Structure V. 10 535 2002.
ISSN: ISSN 0969-2126
PubMed: 11937058
DOI: 10.1016/S0969-2126(02)00738-4
Page generated: Sat Oct 5 11:06:50 2024
ISSN: ISSN 0969-2126
PubMed: 11937058
DOI: 10.1016/S0969-2126(02)00738-4
Last articles
Ca in 5SWICa in 5SVE
Ca in 5SSX
Ca in 5SV0
Ca in 5STD
Ca in 5SSZ
Ca in 5SSY
Ca in 5SIC
Ca in 5SBD
Ca in 5SBE